Peak and Motif analyses in Promoters
gene_x 0 like s 1325 view s
Tags: pipeline, ChIP-seq
-
./1_generate_promoter_sequences.py gencode.v43.annotation.gtf.db
-
./2_find_motifs_in_promoters.py
-
generate peaks_and_motifs_in_promoters_with_LT_peak.xls
peaks_and_motifs_in_promoters_with_LT_peak.xls
#--3.1. generate peak_tss_distances.csv and peak_tss_distances.png python3 plot_peaks.py peaks_NHDF_.bed /home/jhuang/REFs/gencode.v43.annotation.gtf.db /ref/Homo_sapiens/UCSC/hg38/Sequence/WholeGenomeFasta/genome.fa python3 plot_peaks.py peaks_HEK293_.bed /home/jhuang/REFs/gencode.v43.annotation.gtf.db /ref/Homo_sapiens/UCSC/hg38/Sequence/WholeGenomeFasta/genome.fa python3 plot_peaks.py peaks_PFSK-1_.bed /home/jhuang/REFs/gencode.v43.annotation.gtf.db /ref/Homo_sapiens/UCSC/hg38/Sequence/WholeGenomeFasta/genome.fa python3 plot_peaks.py peaks_K331A_.bed /home/jhuang/REFs/gencode.v43.annotation.gtf.db /ref/Homo_sapiens/UCSC/hg38/Sequence/WholeGenomeFasta/genome.fa #--3.2. split data into two dataframe, one is in which PromoterID occurres in filtered_peaks$Closest.Gene..Ensemble.ID., another is the remaining #sed -i 's/[][]//g' motif_counts_positions.txt data <- read.table("motif_counts_positions.txt", sep="\t", header=TRUE, stringsAsFactors=FALSE) # Read the data peaks <- read.table("peak_tss_distances.txt", sep="\t", header=TRUE, stringsAsFactors=FALSE) # Filter the rows filtered_peaks <- subset(peaks, abs(`Distance.to.TSS`) <= 3000) # Print the filtered peaks write.table(filtered_peaks, file="filtered_peak_tss_distances.txt", sep="\t", row.names = FALSE) # Create a new data frame for rows in 'data' where the PromoterID is in filtered_peaks's Closest Gene (Ensemble ID) data_in_filtered <- data[data$PromoterID %in% filtered_peaks$Closest.Gene..Ensemble.ID., ] data_in_filtered$Motif_GRGGC_Center_Positions <- sapply(data_in_filtered$Motif_GRGGC_Center_Positions, toString) data_in_filtered$Motif_GCCYR_Center_Positions <- sapply(data_in_filtered$Motif_GCCYR_Center_Positions, toString) write.table(data_in_filtered, file="motifs_in_promoters_with_LT_peak.txt", sep="\t", row.names = FALSE) # Create another data frame for the remaining rows data_not_in_filtered <- data[!data$PromoterID %in% filtered_peaks$Closest.Gene..Ensemble.ID., ] data_not_in_filtered$Motif_GRGGC_Center_Positions <- sapply(data_not_in_filtered$Motif_GRGGC_Center_Positions, toString) data_not_in_filtered$Motif_GCCYR_Center_Positions <- sapply(data_not_in_filtered$Motif_GCCYR_Center_Positions, toString) write.table(data_not_in_filtered, file="motifs_in_promoters_without_LT_peak.txt", sep="\t", row.names = FALSE) #780 vs 19262 #1707 vs ca. 60000 #sed -i 's/Symbol: //g' motifs_in_promoters_with_LT_peak.txt #sed -i 's/Type: //g' motifs_in_promoters_with_LT_peak.txt #sed -i 's/Symbol: //g' motifs_in_promoters_without_LT_peak.txt #sed -i 's/Type: //g' motifs_in_promoters_without_LT_peak.txt #sed -i 's/"//g' motifs_in_promoters_with_LT_peak.txt #sed -i 's/"//g' motifs_in_promoters_without_LT_peak.txt #--3.3. prepare data for motifs_in_promoters_with_LT_peak.txt data1 <- read.table("motifs_in_promoters_with_LT_peak.txt", sep="\t", header=TRUE, stringsAsFactors=FALSE) data1$Motif_GRGGC_Count <- as.numeric(data1$Motif_GRGGC_Count) data1$Motif_GCCYR_Count <- as.numeric(data1$Motif_GCCYR_Count) data1$Motif_GRGGC_Center_Positions <- strsplit(as.character(data1$Motif_GRGGC_Center_Positions), ",") data1$Motif_GCCYR_Center_Positions <- strsplit(as.character(data1$Motif_GCCYR_Center_Positions), ",") # Create data1 frames for each motif data1_motif1 <- data.frame(PromoterID = rep(data1$PromoterID, data1$Motif_GRGGC_Count), Motif = "Motif GRGGC", Position = unlist(data1$Motif_GRGGC_Center_Positions)) data1_motif2 <- data.frame(PromoterID = rep(data1$PromoterID, data1$Motif_GCCYR_Count), Motif = "Motif GCCYR", Position = unlist(data1$Motif_GCCYR_Center_Positions)) # Combine motif data1 frames data1_positions <- rbind(data1_motif1, data1_motif2) # Get positions as numeric values, excluding NAs motif1_positions_with_LT_peak <- as.numeric(data1_positions$Position[data1_positions$Motif == "Motif GRGGC"]) motif2_positions_with_LT_peak <- as.numeric(data1_positions$Position[data1_positions$Motif == "Motif GCCYR"]) #--3.4. prepare data for motifs_in_promoters_without_LT_peak.txt data2 <- read.table("motifs_in_promoters_without_LT_peak.txt", sep="\t", header=TRUE, stringsAsFactors=FALSE) data2$Motif_GRGGC_Count <- as.numeric(data2$Motif_GRGGC_Count) data2$Motif_GCCYR_Count <- as.numeric(data2$Motif_GCCYR_Count) data2$Motif_GRGGC_Center_Positions <- strsplit(as.character(data2$Motif_GRGGC_Center_Positions), ",") data2$Motif_GCCYR_Center_Positions <- strsplit(as.character(data2$Motif_GCCYR_Center_Positions), ",") # Create data2 frames for each motif data2_motif1 <- data.frame(PromoterID = rep(data2$PromoterID, data2$Motif_GRGGC_Count), Motif = "Motif GRGGC", Position = unlist(data2$Motif_GRGGC_Center_Positions)) data2_motif2 <- data.frame(PromoterID = rep(data2$PromoterID, data2$Motif_GCCYR_Count), Motif = "Motif GCCYR", Position = unlist(data2$Motif_GCCYR_Center_Positions)) # Combine motif data2 frames data2_positions <- rbind(data2_motif1, data2_motif2) # Get positions as numeric values, excluding NAs motif1_positions_without_LT_peak <- as.numeric(data2_positions$Position[data2_positions$Motif == "Motif GRGGC"]) motif2_positions_without_LT_peak <- as.numeric(data2_positions$Position[data2_positions$Motif == "Motif GCCYR"]) #--3.5. Plot Histogram of counts and density of positions # Define a common x-axis limit for all plots x_limit <- max(max(data1$Motif_GRGGC_Count), max(data1$Motif_GCCYR_Count), max(data2$Motif_GRGGC_Count), max(data2$Motif_GCCYR_Count)) # Adjust the number of breaks (nbins) to control the width of histograms nbins <- 30 y_limit1 <- 300 y_limit2 <- 12000 # Plot Histogram of counts with the same scale for x-axis png("Histogram_of_Motif_GRGGC_in_promoters_with_LT_peak.png", width=800, height=800) hist(data1$Motif_GRGGC_Count, main = "Histogram of Motif GRGGC in promoters with LT peak", xlab = "Counts", xlim = c(0, x_limit), ylim = c(0, y_limit1), breaks = 20) dev.off() png("Histogram_of_Motif_GRGGC_in_promoters_without_LT_peak.png", width=800, height=800) hist(data2$Motif_GRGGC_Count, main = "Histogram of Motif GRGGC in promoters without LT peak", xlab = "Counts", xlim = c(0, x_limit), ylim = c(0, y_limit2), breaks = nbins) dev.off() png("Histogram_of_Motif_GCCYR_in_promoters_with_LT_peak.png", width=800, height=800) hist(data1$Motif_GCCYR_Count, main = "Histogram of Motif GCCYR in promoters with LT peak", xlab = "Counts", xlim = c(0, x_limit), ylim = c(0, y_limit1), breaks = nbins) dev.off() png("Histogram_of_Motif_GCCYR_in_promoters_without_LT_peak.png", width=800, height=800) hist(data2$Motif_GCCYR_Count, main = "Histogram of Motif GCCYR in promoters without LT peak", xlab = "Counts", xlim = c(0, x_limit), ylim = c(0, y_limit2), breaks = nbins) dev.off() # Set up the layout for the plots png("Histogram_of_Motifs.png", width = 1000, height = 1000) par(mfrow = c(2, 2)) hist(data1$Motif_GRGGC_Count, main = "Motif GRGGC Counts with LT peak", xlab = "Counts", xlim = c(0, x_limit), ylim = c(0, y_limit1), breaks = 20) hist(data2$Motif_GRGGC_Count, main = "Motif GRGGC Counts without LT peak", xlab = "Counts", xlim = c(0, x_limit), ylim = c(0, y_limit2), breaks = nbins) hist(data1$Motif_GCCYR_Count, main = "Motif GCCYR Counts with LT peak", xlab = "Counts", xlim = c(0, x_limit), ylim = c(0, y_limit1), breaks = nbins) hist(data2$Motif_GCCYR_Count, main = "Motif GCCYR Counts without LT peak", xlab = "Counts", xlim = c(0, x_limit), ylim = c(0, y_limit2), breaks = nbins) dev.off() # Set the ylim value ylim <- c(0.0001, 0.0003) png("Density_of_Motif_GRGGC_in_promoters_with_LT_peak.png", width=800, height=800) plot(density(motif1_positions_with_LT_peak, na.rm = TRUE), main="Density of GRGGC in promoters with LT peak", xlab="Position", ylim = ylim) dev.off() png("Density_of_Motif_GRGGC_in_promoters_without_LT_peak.png", width=800, height=800) plot(density(motif1_positions_without_LT_peak, na.rm = TRUE), main="Density of GRGGC in promoters without LT peak", xlab="Position", ylim = ylim) dev.off() png("Density_of_Motif_GCCYR_in_promoters_with_LT_peak.png", width=800, height=800) plot(density(motif2_positions_with_LT_peak, na.rm = TRUE), main="Density of GCCYR in promoters with LT peak", xlab="Position", ylim = ylim) dev.off() png("Density_of_Motif_GCCYR_in_promoters_without_LT_peak.png", width=800, height=800) plot(density(motif2_positions_without_LT_peak, na.rm = TRUE), main="Density of GCCYR in promoters without LT peak", xlab="Position", ylim = ylim) dev.off() png("Density_of_Motifs.png", width = 1000, height = 1000) par(mfrow = c(2, 2)) plot(density(motif1_positions_with_LT_peak, na.rm = TRUE), main = "Density of GRGGC in promoters with LT peak", xlab = "Position", ylim = ylim) plot(density(motif1_positions_without_LT_peak, na.rm = TRUE), main = "Density of GRGGC in promoters without LT peak", xlab = "Position", ylim = ylim) plot(density(motif2_positions_with_LT_peak, na.rm = TRUE), main = "Density of GCCYR in promoters with LT peak", xlab = "Position", ylim = ylim) plot(density(motif2_positions_without_LT_peak, na.rm = TRUE), main = "Density of GCCYR in promoters without LT peak", xlab = "Position", ylim = ylim) dev.off() #--3.6. assign each promoter to a prefined class according to Motifs_Count and Avg_Distance_To_TSS_of_Motifs library(dplyr) data <- read.table("motifs_in_promoters_with_LT_peak.txt", sep="\t", header=TRUE, stringsAsFactors=FALSE) data$Motif_GRGGC_Count <- as.numeric(data$Motif_GRGGC_Count) data$Motif_GCCYR_Count <- as.numeric(data$Motif_GCCYR_Count) # Add a column for the sum of Motif_GRGGC_Count and Motif_GCCYR_Count data <- data %>% mutate(Motifs_Count = Motif_GRGGC_Count + Motif_GCCYR_Count) # Calculate absolute average Motif_GRGGC distance to TSS data$Avg_Distance_To_TSS_of_Motif_GRGGC <- sapply(strsplit(data$Motif_GRGGC_Center_Positions, ", "), function(x) round(mean(as.numeric(x), na.rm = TRUE))) # Calculate absolute average Motif_GCCYR distance to TSS data$Avg_Distance_To_TSS_of_Motif_GCCYR <- sapply(strsplit(data$Motif_GCCYR_Center_Positions, ", "), function(x) round(mean(as.numeric(x), na.rm = TRUE))) # Revised function to calculate the mean of two strings of comma-separated numbers mean_of_strings <- function(str1, str2) { # Convert strings of numbers to numeric vectors vec1 <- as.numeric(strsplit(str1, split = ",")[[1]]) vec2 <- as.numeric(strsplit(str2, split = ",")[[1]]) # Handle cases where vec1 or vec2 is NULL (i.e., when str1 or str2 is an empty string) if (is.null(vec1)) { vec1 <- numeric(0) } if (is.null(vec2)) { vec2 <- numeric(0) } # Compute the mean of the two vectors return(round(mean(c(vec1, vec2), na.rm = TRUE))) } # Apply the function to each row of the dataframe data$Avg_Distance_To_TSS_of_Motifs <- mapply(mean_of_strings, data$Motif_GRGGC_Center_Positions, data$Motif_GCCYR_Center_Positions) # Assign classes based on the given rules data <- data %>% mutate( Class_based_Motif_GRGGC = case_when( Motif_GRGGC_Count < 50 & abs(Avg_Distance_To_TSS_of_Motif_GRGGC) <= 500 ~ "Class 1", Motif_GRGGC_Count < 50 & abs(Avg_Distance_To_TSS_of_Motif_GRGGC) > 500 & abs(Avg_Distance_To_TSS_of_Motif_GRGGC) <= 1000 ~ "Class 2", Motif_GRGGC_Count < 50 & abs(Avg_Distance_To_TSS_of_Motif_GRGGC) > 1000 ~ "Class 3", Motif_GRGGC_Count >= 50 & Motif_GRGGC_Count < 100 & abs(Avg_Distance_To_TSS_of_Motif_GRGGC) <= 500 ~ "Class 4", Motif_GRGGC_Count >= 50 & Motif_GRGGC_Count < 100 & abs(Avg_Distance_To_TSS_of_Motif_GRGGC) > 500 & abs(Avg_Distance_To_TSS_of_Motif_GRGGC) <= 1000 ~ "Class 5", Motif_GRGGC_Count >= 50 & Motif_GRGGC_Count < 100 & abs(Avg_Distance_To_TSS_of_Motif_GRGGC) > 1000 ~ "Class 6", Motif_GRGGC_Count >= 100 & Motif_GRGGC_Count < 150 & abs(Avg_Distance_To_TSS_of_Motif_GRGGC) <= 500 ~ "Class 7", Motif_GRGGC_Count >= 100 & Motif_GRGGC_Count < 150 & abs(Avg_Distance_To_TSS_of_Motif_GRGGC) > 500 & abs(Avg_Distance_To_TSS_of_Motif_GRGGC) <= 1000 ~ "Class 8", Motif_GRGGC_Count >= 100 & Motif_GRGGC_Count < 150 & abs(Avg_Distance_To_TSS_of_Motif_GRGGC) > 1000 ~ "Class 9" ) ) data <- data %>% mutate( Class_based_Motif_GCCYR = case_when( Motif_GCCYR_Count < 50 & abs(Avg_Distance_To_TSS_of_Motif_GCCYR) <= 500 ~ "Class 1", Motif_GCCYR_Count < 50 & abs(Avg_Distance_To_TSS_of_Motif_GCCYR) > 500 & abs(Avg_Distance_To_TSS_of_Motif_GCCYR) <= 1000 ~ "Class 2", Motif_GCCYR_Count < 50 & abs(Avg_Distance_To_TSS_of_Motif_GCCYR) > 1000 ~ "Class 3", Motif_GCCYR_Count >= 50 & Motif_GCCYR_Count < 100 & abs(Avg_Distance_To_TSS_of_Motif_GCCYR) <= 500 ~ "Class 4", Motif_GCCYR_Count >= 50 & Motif_GCCYR_Count < 100 & abs(Avg_Distance_To_TSS_of_Motif_GCCYR) > 500 & abs(Avg_Distance_To_TSS_of_Motif_GCCYR) <= 1000 ~ "Class 5", Motif_GCCYR_Count >= 50 & Motif_GCCYR_Count < 100 & abs(Avg_Distance_To_TSS_of_Motif_GCCYR) > 1000 ~ "Class 6", Motif_GCCYR_Count >= 100 & Motif_GCCYR_Count < 150 & abs(Avg_Distance_To_TSS_of_Motif_GCCYR) <= 500 ~ "Class 7", Motif_GCCYR_Count >= 100 & Motif_GCCYR_Count < 150 & abs(Avg_Distance_To_TSS_of_Motif_GCCYR) > 500 & abs(Avg_Distance_To_TSS_of_Motif_GCCYR) <= 1000 ~ "Class 8", Motif_GCCYR_Count >= 100 & Motif_GCCYR_Count < 150 & abs(Avg_Distance_To_TSS_of_Motif_GCCYR) > 1000 ~ "Class 9" ) ) data <- data %>% mutate( Class_based_Motifs = case_when( Motifs_Count < 50 & abs(Avg_Distance_To_TSS_of_Motifs) <= 500 ~ "Class 1", Motifs_Count < 50 & abs(Avg_Distance_To_TSS_of_Motifs) > 500 & abs(Avg_Distance_To_TSS_of_Motifs) <= 1000 ~ "Class 2", Motifs_Count < 50 & abs(Avg_Distance_To_TSS_of_Motifs) > 1000 ~ "Class 3", Motifs_Count >= 50 & Motifs_Count < 100 & abs(Avg_Distance_To_TSS_of_Motifs) <= 500 ~ "Class 4", Motifs_Count >= 50 & Motifs_Count < 100 & abs(Avg_Distance_To_TSS_of_Motifs) > 500 & abs(Avg_Distance_To_TSS_of_Motifs) <= 1000 ~ "Class 5", Motifs_Count >= 50 & Motifs_Count < 100 & abs(Avg_Distance_To_TSS_of_Motifs) > 1000 ~ "Class 6", Motifs_Count >= 100 & Motifs_Count < 150 & abs(Avg_Distance_To_TSS_of_Motifs) <= 500 ~ "Class 7", Motifs_Count >= 100 & Motifs_Count < 150 & abs(Avg_Distance_To_TSS_of_Motifs) > 500 & abs(Avg_Distance_To_TSS_of_Motifs) <= 1000 ~ "Class 8", Motifs_Count >= 100 & Motifs_Count < 150 & abs(Avg_Distance_To_TSS_of_Motifs) > 1000 ~ "Class 9" ) ) # Define the desired column order desired_columns <- c("PromoterID","Gene_Symbol","Gene_Type","Motif_GRGGC_Count","Motif_GRGGC_Center_Positions","Avg_Distance_To_TSS_of_Motif_GRGGC","Class_based_Motif_GRGGC", "Motif_GCCYR_Count","Motif_GCCYR_Center_Positions","Avg_Distance_To_TSS_of_Motif_GCCYR","Class_based_Motif_GCCYR", "Motifs_Count","Avg_Distance_To_TSS_of_Motifs","Class_based_Motifs") # Reorder the columns data <- select(data, all_of(desired_columns)) #sed -i 's/Motifs/Motifs_GRGGC+GCCYR/g' motifs_in_promoters_with_LT_peak_Category.txt write.table(data, file="motifs_in_promoters_with_LT_peak_Category.txt", sep="\t", row.names = FALSE) #--3.7. add peak information into motif table ./add_peaks_to_motifs.py #~/Tools/csv2xls-0.4/csv_to_xls.py _peaks_and_motifs_in_promoters_with_LT_peak.txt peak_tss_distances.txt -d$'\t' -o peaks_and_motifs_in_promoters_with_LT_peak.xls; #rename the name of sheet1 to "peaks_and_motifs_in_promoters_with_LT_peak" and "all_LT_peaks_NHDF" -
plot_peaks.py (updated code)
-
plot_peaks.py (updated code)
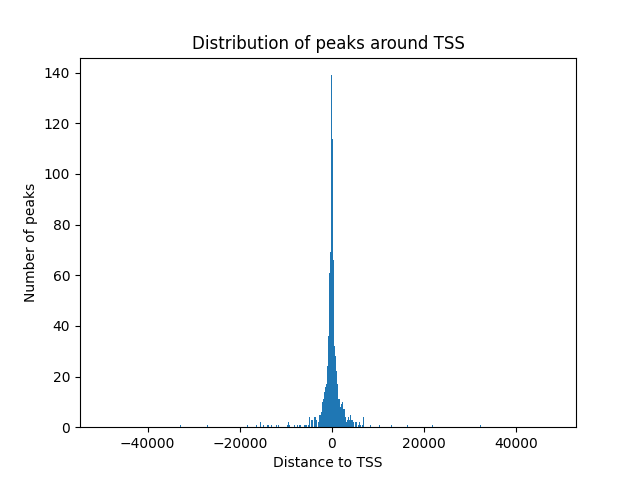
#python3 plot_peaks.py peaks_NHDF_.bed /home/jhuang/REFs/gencode.v43.annotation.gtf.db #mv peak_tss_distances.xlsx peak_tss_distances_NHDF.xlsx #mv peak_tss_distances.txt peak_tss_distances_NHDF.txt #mv peak_distribution.csv peak_distribution_NHDF.csv #mv peak_distribution.png peak_distribution_NHDF.png #python3 plot_peaks.py peaks_HEK293_.bed /home/jhuang/REFs/gencode.v43.annotation.gtf.db #mv peak_tss_distances.xlsx peak_tss_distances_HEK293.xlsx #mv peak_tss_distances.txt peak_tss_distances_HEK293.txt #mv peak_distribution.csv peak_distribution_HEK293.csv #mv peak_distribution.png peak_distribution_HEK293.png #python3 plot_peaks.py peaks_PFSK-1_.bed /home/jhuang/REFs/gencode.v43.annotation.gtf.db #mv peak_tss_distances.xlsx peak_tss_distances_PFSK-1.xlsx #mv peak_tss_distances.txt peak_tss_distances_PFSK-1.txt #mv peak_distribution.csv peak_distribution_PFSK-1.csv #mv peak_distribution.png peak_distribution_PFSK-1.png import pprint import argparse import matplotlib.pyplot as plt import pandas as pd import gffutils import numpy as np def plot_peak_distribution(peaks_file, gencode_db): db = gffutils.FeatureDB(gencode_db, keep_order=True) peaks = pd.read_table(peaks_file, header=None) peaks.columns = ['chrom', 'start', 'end', 'name', 'score'] tss_positions = [] tss_genes = [] for gene in db.features_of_type('gene'): if gene.strand == '+': tss_positions.append(gene.start) else: tss_positions.append(gene.end) tss_genes.append(gene) peak_tss_distances = [] peak_names = [] closest_genes = [] closest_gene_names = [] closest_strands = [] closest_gene_types = [] # New list for gene_type for _, peak in peaks.iterrows(): peak_center = (peak['start'] + peak['end']) // 2 closest_tss_index = np.argmin([abs(tss - peak_center) for tss in tss_positions]) distance_to_tss = peak_center - tss_positions[closest_tss_index] peak_tss_distances.append(distance_to_tss) peak_names.append(peak['name']) closest_genes.append(tss_genes[closest_tss_index].id) if 'gene_name' in tss_genes[closest_tss_index].attributes: closest_gene_name = tss_genes[closest_tss_index].attributes['gene_name'][0] else: closest_gene_name = 'N/A' # Set a default value if 'gene_name' attribute is not present closest_gene_names.append(closest_gene_name) closest_strands.append(tss_genes[closest_tss_index].strand) if 'gene_type' in tss_genes[closest_tss_index].attributes: closest_gene_type = tss_genes[closest_tss_index].attributes['gene_type'][0] else: closest_gene_type = 'N/A' # Set a default value if 'gene_type' attribute is not present closest_gene_types.append(closest_gene_type) df = pd.DataFrame({ 'Peak Name': peak_names, 'Distance to TSS': peak_tss_distances, 'Closest Gene (Ensemble ID)': closest_genes, 'Closest Gene (Gene name)': closest_gene_names, 'Gene Strand': closest_strands, 'Gene Type': closest_gene_types # Add new column for gene_type }) df.to_excel('peak_tss_distances.xlsx', index=False) df.to_csv('peak_tss_distances.txt', sep='\t', index=False) # Save as tab-separated text file bins_no=600 #1000 for nHDF, 600 for HEK293 and PFSK-1 counts, bin_edges = np.histogram(peak_tss_distances, bins=bins_no) bin_centers = (bin_edges[:-1] + bin_edges[1:]) / 2 hist_df = pd.DataFrame({'Bin Center': bin_centers, 'Count': counts}) hist_df.to_csv('peak_distribution.csv', index=False) total_peaks = hist_df['Count'].sum() with open('peak_distribution.csv', 'a') as f: f.write(f'\nTotal number of peaks: {total_peaks}') # Set the figure background to be transparent plt.gcf().set_facecolor('none') plt.gcf().patch.set_alpha(0.0) plt.hist(peak_tss_distances, bins=bins_no) plt.xlabel('Distance to TSS', fontsize=17) plt.ylabel('Number of peaks', fontsize=17) plt.title('') #plt.title('Distribution of peaks around TSS', fontsize=12) # Set tick label font sizes plt.xticks(fontsize=14) plt.yticks(fontsize=14) # Adjust the space on the left of the figure plt.gcf().subplots_adjust(left=0.15) # Set x-axis range plt.xlim(-8000, 8000) plt.savefig('peak_distribution.png', dpi=300, bbox_inches='tight') plt.show() if __name__ == "__main__": parser = argparse.ArgumentParser(description='Plot peak distribution around TSS.') parser.add_argument('peaks_file', type=str, help='Path to the peaks.bed file') parser.add_argument('gencode_db', type=str, help='Path to the gencode.v43.annotation.db file') args = parser.parse_args() plot_peak_distribution(args.peaks_file, args.gencode_db)
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Kraken2 Installation and Usage Guide
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Should the inputs for GSVA be normalized or raw?
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- Risks of Rebooting into Rescue Mode
- 足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Peak calling using homer combining sicer and macs2
How to use H3K27ac, H3K4me1, and RNA-seq to identify enhancers and their target genes?