PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
gene_x 0 like s 2581 view s
Tags: metagenomics, 16S
How to run the software package PiCRUST2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States version 2), and to visualize its output data using STAMP (Statistical Analysis of Metagenomic Profiles) and ALDEx2 (Analysis of Differential Abundance taking sample Variation into Account version 2) for Functional Prediction and Pathway Analysis in Metagenomics.
-
Environment Setup: It sets up a Conda environment named picrust2, using the conda create command and then activates this environment using conda activate picrust2.
#https://github.com/picrust/picrust2/wiki/PICRUSt2-Tutorial-(v2.2.0-beta)#minimum-requirements-to-run-full-tutorial conda create -n picrust2 -c bioconda -c conda-forge picrust2 #=2.2.0_b conda activate picrust2 -
Data Preparation: The script creates a new directory called picrust2_out, then enters it using mkdir and cd commands. It then identifies input files that are needed for the analysis: metadata.tsv, seqs.fna, table.biom. The biom commands are used to inspect and convert the BIOM format files.
mkdir picrust2_out cd picrust2_out # Identifying input data # Note: Replace the paths and filenames with your actual data if different # metadata.tsv == ../map_corrected.txt # seqs.fna == ../clustering/seqs.fna # table.biom == ../core_diversity_e42369/table_even42369.biom # Inspect and convert the BIOM format files biom head -i ../core_diversity_e42369/table_even42369.biom biom summarize-table -i ../core_diversity_e42369/table_even42369.biom biom convert -i ../core_diversity_e42369/table_even42369.biom -o table_even42369.tsv --to-tsv -
Running PiCRUST2: The place_seqs.py command aligns the input sequences to a reference tree. The hsp.py commands generate hidden state prediction for multiple functional categories.
#insert reads into reference tree using EPA-NG cp ../clustering/rep_set.fna ./ grep ">" rep_set.fna | wc -l #44238 vim table_even42369.tsv #40596-2 samtools faidx rep_set.fna cut -f1-1 table_even42369.tsv > table_even42369.id #manually modify table_even42369.id by replacing "\n" with " >> seqs.fna\nsamtools faidx rep_set.fna " run table_even42369.id rm -rf intermediate/ place_seqs.py -s seqs.fna -o out.tre -p 4 --intermediate intermediate/place_seqs #castor: Efficient Phylogenetics on Large Trees #https://github.com/picrust/picrust2/wiki/Hidden-state-prediction hsp.py -i 16S -t out.tre -o 16S_predicted_and_nsti.tsv.gz -p 15 -n hsp.py -i COG -t out.tre -o COG_predicted.tsv.gz -p 15 hsp.py -i PFAM -t out.tre -o PFAM_predicted.tsv.gz -p 15 hsp.py -i KO -t out.tre -o KO_predicted.tsv.gz -p 15 hsp.py -i EC -t out.tre -o EC_predicted.tsv.gz -p 15 hsp.py -i TIGRFAM -t out.tre -o TIGRFAM_predicted.tsv.gz -p 15 hsp.py -i PHENO -t out.tre -o PHENO_predicted.tsv.gz -p 15In this table the predicted copy number of all Enzyme Classification (EC) numbers is shown for each ASV. The NSTI values per ASV are not in this table since we did not specify the -n option. EC numbers are a type of gene family defined based on the chemical reactions they catalyze. For instance, EC:1.1.1.1 corresponds to alcohol dehydrogenase. In this tutorial we are focusing on EC numbers since they can be used to infer MetaCyc pathway levels (see below).
zless -S EC_predicted.tsv.gz sequence EC:1.1.1.1 EC:1.1.1.10 EC:1.1.1.100 ... 20e568023c10eaac834f1c110aacea18 2 0 3 ... 23fe12a325dfefcdb23447f43b6b896e 0 0 1 ... 288c8176059111c4c7fdfb0cd5afce64 1 0 1 ... ... ##Why import the tsv file to MyData? #MyData <- read.csv(file="./COG_predicted.tsv", header=TRUE, sep="\t", row.names=1) #6806 4598 e.g. COG5665 #MyData <- read.csv(file="./PFAM_predicted.tsv", header=TRUE, sep="\t", row.names=1) #6806 11089 e.g. PF17225 #MyData <- read.csv(file="./KO_predicted.tsv", header=TRUE, sep="\t", row.names=1) #6806 10543 e.g. K19791 #MyData <- read.csv(file="./EC_predicted.tsv", header=TRUE, sep="\t", row.names=1) #6806 2913 e.g. EC.6.6.1.2 #MyData <- read.csv(file="./16S_predicted.tsv", header=TRUE, sep="\t", row.names=1) #6806 1 e.g. X16S_rRNA_Count #MyData <- read.csv(file="./TIGRFAM_predicted.tsv", header=TRUE, sep="\t", row.names=1) #6806 4287 e.g. TIGR04571 #MyData <- read.csv(file="./PHENO_predicted.tsv", header=TRUE, sep="\t", row.names=1) #6806 41 e.g. Use_of_nitrate_as_electron_acceptor, Xylose_utilizing -
The metagenome_pipeline.py commands perform metagenomic prediction for several functional categories. Predicted gene families weighted by the relative abundance of ASVs in their community. In other words, we are interested in inferring the metagenomes of the communities.
#Generate metagenome predictions using EC numbers https://en.wikipedia.org/wiki/List_of_enzymes#Category:EC_1.1_(act_on_the_CH-OH_group_of_donors) metagenome_pipeline.py -i ../core_diversity_e42369/table_even42369.biom -m 16S_predicted_and_nsti.tsv.gz -f COG_predicted.tsv.gz -o COG_metagenome_out --strat_out metagenome_pipeline.py -i ../core_diversity_e42369/table_even42369.biom -m 16S_predicted_and_nsti.tsv.gz -f EC_predicted.tsv.gz -o EC_metagenome_out --strat_out metagenome_pipeline.py -i ../core_diversity_e42369/table_even42369.biom -m 16S_predicted_and_nsti.tsv.gz -f KO_predicted.tsv.gz -o KO_metagenome_out --strat_out metagenome_pipeline.py -i ../core_diversity_e42369/table_even42369.biom -m 16S_predicted_and_nsti.tsv.gz -f PFAM_predicted.tsv.gz -o PFAM_metagenome_out --strat_out metagenome_pipeline.py -i ../core_diversity_e42369/table_even42369.biom -m 16S_predicted_and_nsti.tsv.gz -f TIGRFAM_predicted.tsv.gz -o TIGRFAM_metagenome_out --strat_out -
Pathway-level inference: By default this script infers MetaCyc pathway abundances based on EC number abundances, although different gene families and pathways can also be optionally specified. This script performs a number of steps by default, which are based on the approach implemented in HUMAnN2:
- Regroups EC numbers to MetaCyc reactions.
- Infers which MetaCyc pathways are present based on these reactions with MinPath.
- Calculates and returns the abundance of pathways identified as present.
#pathway_pipeline.py -i EC_metagenome_out/pred_metagenome_contrib.tsv.gz -o pathways_out -p 15 #Note that the path of map files is under /home/jhuang/anaconda3/envs/picrust2/lib/python3.6/site-packages/picrust2/default_files/pathway_mapfiles (picrust2) pathway_pipeline.py -i COG_metagenome_out/pred_metagenome_contrib.tsv.gz -o KEGG_pathways_out -p 15 --no_regroup --map /home/jhuang/anaconda3/envs/picrust2/lib/python3.6/site-packages/picrust2/default_files/pathway_mapfiles/KEGG_pathways_to_KO.tsv #Mapping predicted KO abundances to legacy KEGG pathways (with stratified output that represents contributions to community-wide abundances): (picrust2) pathway_pipeline.py -i KO_metagenome_out/pred_metagenome_strat.tsv.gz -o KEGG_pathways_out --no_regroup --map /home/jhuang/anaconda3/envs/picrust2/lib/python3.6/site-packages/picrust2/default_files/pathway_mapfiles/KEGG_pathways_to_KO.tsv #Map EC numbers to MetaCyc pathways and get stratified output corresponding to contribution of predicted gene family abundances within each predicted genome: #BUG: CANNOT FINISH in 1 day! (picrust2) pathway_pipeline.py -i EC_metagenome_out/pred_metagenome_unstrat.tsv.gz -o pathways_out_per_seq --per_sequence_contrib --per_sequence_abun EC_metagenome_out/seqtab_norm.tsv.gz --per_sequence_function EC_predicted.tsv.gz (picrust2) pathway_pipeline.py -i EC_metagenome_out/pred_metagenome_unstrat.tsv.gz -o pathways_out -p 6
-
Add functional descriptions: Finally, it can be useful to have a description of each functional id in the output abundance tables. The below commands will add these descriptions as new column in gene family and pathway abundance tables
#--6.1. Add descriptions in gene family tables add_descriptions.py -i COG_metagenome_out/pred_metagenome_unstrat.tsv.gz -m COG -o COG_metagenome_out/pred_metagenome_unstrat_descrip.tsv.gz add_descriptions.py -i EC_metagenome_out/pred_metagenome_unstrat.tsv.gz -m EC -o EC_metagenome_out/pred_metagenome_unstrat_descrip.tsv.gz add_descriptions.py -i KO_metagenome_out/pred_metagenome_unstrat.tsv.gz -m KO -o KO_metagenome_out/pred_metagenome_unstrat_descrip.tsv.gz # EC and METACYC is a pair, EC for gene_annotation and METACYC for pathway_annotation add_descriptions.py -i PFAM_metagenome_out/pred_metagenome_unstrat.tsv.gz -m PFAM -o PFAM_metagenome_out/pred_metagenome_unstrat_descrip.tsv.gz add_descriptions.py -i TIGRFAM_metagenome_out/pred_metagenome_unstrat.tsv.gz -m TIGRFAM -o TIGRFAM_metagenome_out/pred_metagenome_unstrat_descrip.tsv.gz #--6.2. Add descriptions in pathway abundance tables add_descriptions.py -i pathways_out/path_abun_unstrat.tsv.gz -m METACYC -o pathways_out/path_abun_unstrat_descrip.tsv.gz #Error - no rows remain after regrouping input table. The default pathway and regroup mapfiles are meant for EC numbers. Note that KEGG pathways are not supported since KEGG is a closed-source database, but you can input custom pathway mapfiles if you have access. If you are using a custom function database did you mean to set the --no-regroup flag and/or change the default pathways mapfile used? #If ERROR --> USE the METACYC for downstream analyses!!! add_descriptions.py -i pathways_out/path_abun_unstrat.tsv.gz -o KEGG_pathways_out/path_abun_unstrat_descrip.tsv.gz --custom_map_table /home/jhuang/anaconda3/envs/picrust2/lib/python3.6/site-packages/picrust2/default_files/description_mapfiles/KEGG_pathways_info.tsv.gz -
Visualization
#7.1 install and open STAMP #https://github.com/picrust/picrust2/wiki/STAMP-example #install and open STAMP conda deactivate conda install -c bioconda stamp #sudo pip install pyqi #sudo apt-get install libblas-dev liblapack-dev gfortran #sudo apt-get install freetype* python-pip python-dev python-numpy python-scipy python-matplotlib #sudo pip install STAMP #conda install -c bioconda stamp conda create -n stamp -c bioconda/label/cf201901 stamp brew install pyqt #DEBUG the environment conda install pyqt=4 #conda install icu=56 (stamp) jhuang@hamburg:~$ STAMP #7.2 unzip path_abun_unstrat_descrip.tsv.gz gunzip path_abun_unstrat_descrip.tsv.gz #7.3 prepare metadata.tsv from map_corrected.txt vim metadata.tsv #SampleID Genotype Description #S1 Before_non-reducers IDFrancesco1 #S2 After_non-reducers IDFrancesco2 #S3 Before_Reducers IDFrancesco4 #S4 After_Reducers IDFrancesco5 cut -d$'\t' -f1 map_corrected.txt > 1 cut -d$'\t' -f5 map_corrected.txt > 5 cut -d$'\t' -f6 map_corrected.txt > 6 paste -d$'\t' 1 5 > 1_5 paste -d$'\t' 1_5 6 > metadata.tsv #SampleID --> SampleID SampleID Facility Genotype 100CHE6KO PaloAlto KO 101CHE6WT PaloAlto WT #7.4(optional) use ALDEx2 rather than STAMP: https://bioconductor.org/packages/release/bioc/html/ALDEx2.html -
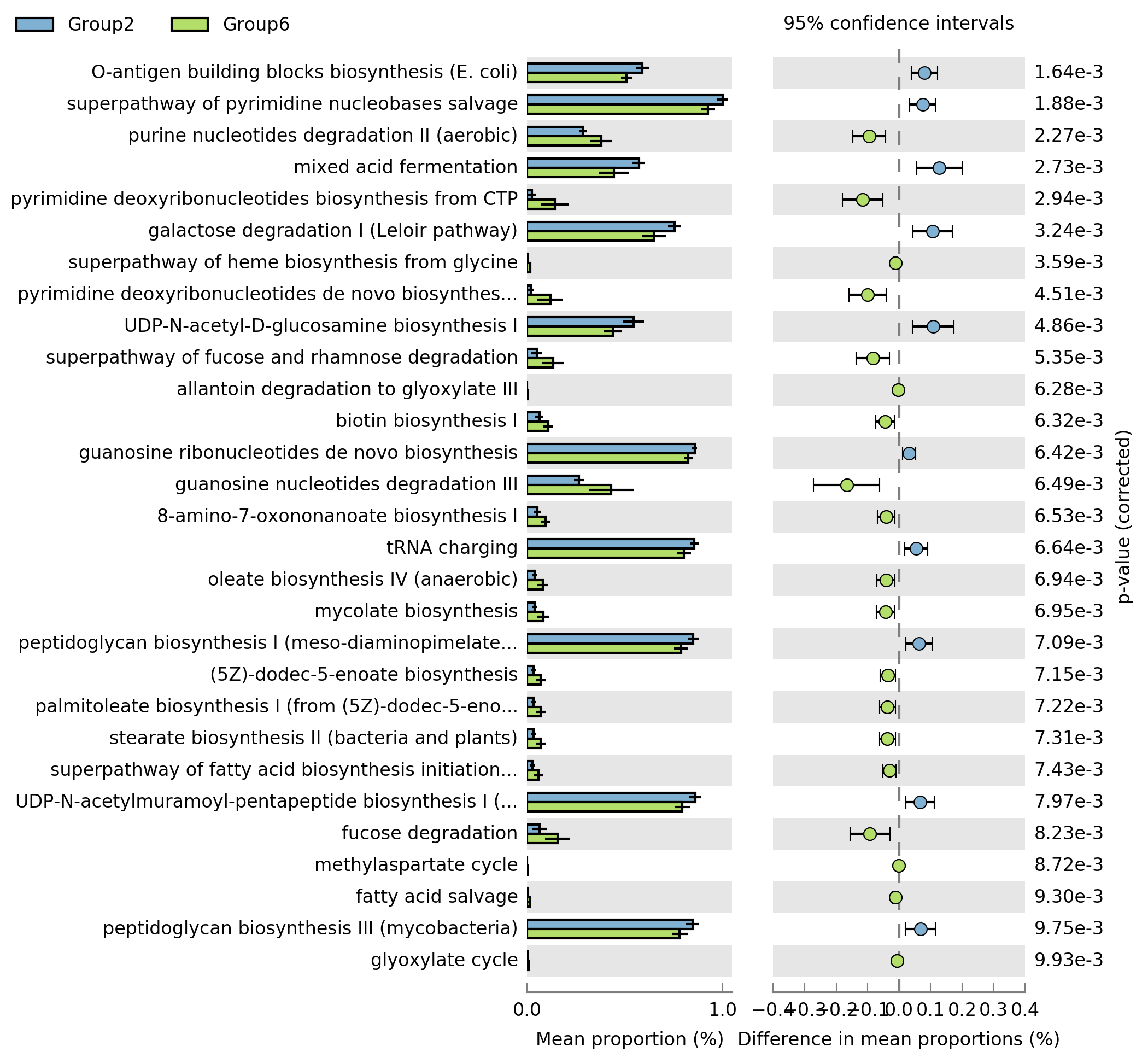
Explanation of the generated plot from Step 7: Extended error bar.
The difference in mean proportions is a statistical measurement that is often used in comparing the proportions of a certain outcome between two groups.
Here's a simple example to explain the concept:
Imagine you conducted a survey on two groups of people, Group A and Group B, asking whether they like a specific brand of chocolate. In Group A, 70 out of 100 people said yes (proportion = 0.7). In Group B, 80 out of 100 people said yes (proportion = 0.8). The difference in proportions is 0.8 - 0.7 = 0.1. This means that in your sample, the proportion of people who like the specific brand of chocolate is 10% higher in Group B compared to Group A.
Statistically speaking, we often want to know whether this difference is significant (i.e., is it likely to be due to chance, or is there a real difference between the groups?). We can use a statistical test, such as a two-proportion z-test, to answer this question.
It's important to note that the difference in proportions is sensitive to the size of your sample. If you have very large groups, even a small difference in proportions can be statistically significant. If you have small groups, only a large difference will be statistically significant.
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Kraken2 Installation and Usage Guide
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Should the inputs for GSVA be normalized or raw?
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- pheatmap vs heatmap.2
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- Risks of Rebooting into Rescue Mode
- 足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
QIIME + Phyloseq + MicrobiotaProcess (v1)
Clinical metagenomics [Talks for Shenzhen and so on]