4sU-tagging, ribosome profiling and ATAC-Seq
gene_x 0 like s 1034 view s
Tags: research
https://www.virologie.uni-wuerzburg.de/virologie/ags-virologie/ag-doelken/research/
-
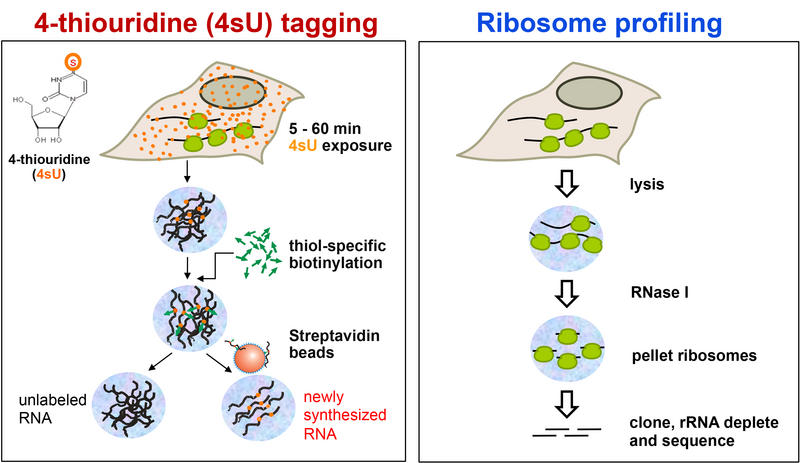
Principle of 4sU-tagging: 4-thiouridine (4sU) is added to cell culture medium to start metabolic labelling of newly transcribed (nascent) RNA. 5 to 60 min later total RNA is isolated. Following thiol-specific biotinylation, total RNA is separated into labelled nascent and unlabelled pre-existing RNA. All three RNA fractions are suitable for down-stream analyses including qRT-PCR, microarrays and RNA-seq.
-
Principle of ribosome profiling: Following a mild cell lysis, a stringent RNAse digest is performed. The short fragment of RNA molecules actively being translated at the time of cell lysis is protected from RNAse treatment within the ribosome and can then be recovered, cloned and sequenced. Thereby, a detailed picture of translational activity at the time of cell lysis is obtained
-
Chromatin accessibility profiling by ATAC-Seq
https://www.nature.com/articles/s41596-022-00692-9 https://www.illumina.com/techniques/multiomics/epigenetics/atac-seq-chromatin-accessibility.html
-
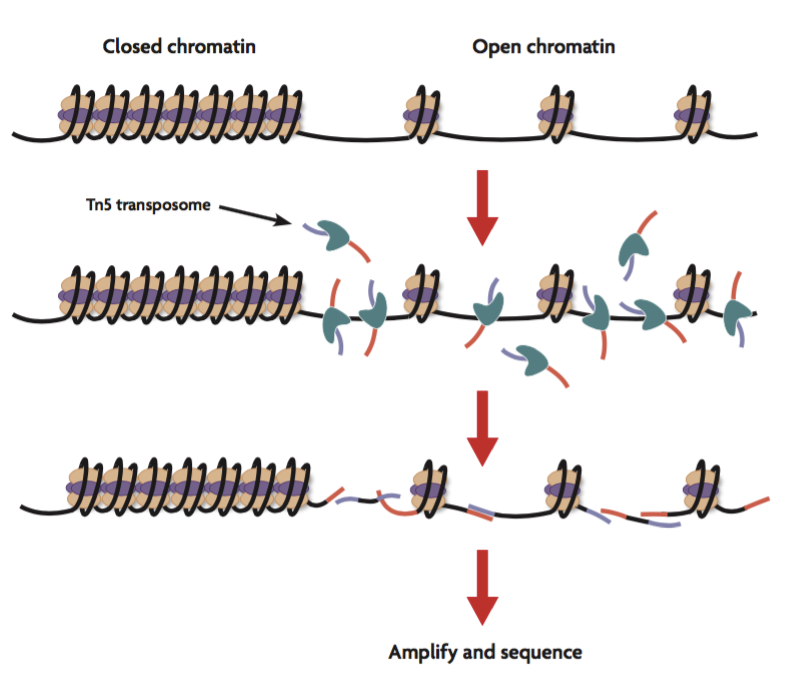
The assay for transposase-accessible chromatin with sequencing (ATAC-Seq) is a popular method for determining chromatin accessibility across the genome. By sequencing regions of open chromatin, ATAC-Seq can help you uncover how chromatin packaging and other factors affect gene expression. ATAC-Seq does not require prior knowledge of regulatory elements, making it a powerful epigenetic discovery tool. It has been used to better understand chromatin accessibility, transcription factor binding, and gene regulation in complex diseases, embryonic development, T-cell activation, and cancer. ATAC-Seq can be performed on bulk cell populations or on single cells at high resolution.
-
How Does ATAC-Seq Work? In ATAC-Seq, genomic DNA is exposed to Tn5, a highly active transposase. Tn5 simultaneously fragments DNA, preferentially inserts into open chromatin sites, and adds sequencing primers (a process known as tagmentation). The sequenced DNA identifies the open chromatin and data analysis can provide insight into gene regulation. Additionally, ATAC-Seq can be combined with other methods, such as RNA sequencing, for a multiomic approach to studying gene expression. Subsequent experiments often include ChIP-Seq, Methyl-Seq, or Hi-C-Seq to further characterize forms of epigenetic regulation.
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Kraken2 Installation and Usage Guide
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Should the inputs for GSVA be normalized or raw?
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- Risks of Rebooting into Rescue Mode
- 足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
基因组与表型特性研究:一株药物敏感的鲍曼不动杆菌显示出增强的毒力相关特性和应激耐受性