The structural proteome of eukaryotic viruses
gene_x 0 like s 1336 view s
Tags: research, protein
Original text please find under https://www.biorxiv.org/content/10.1101/2024.01.22.576744v1.full
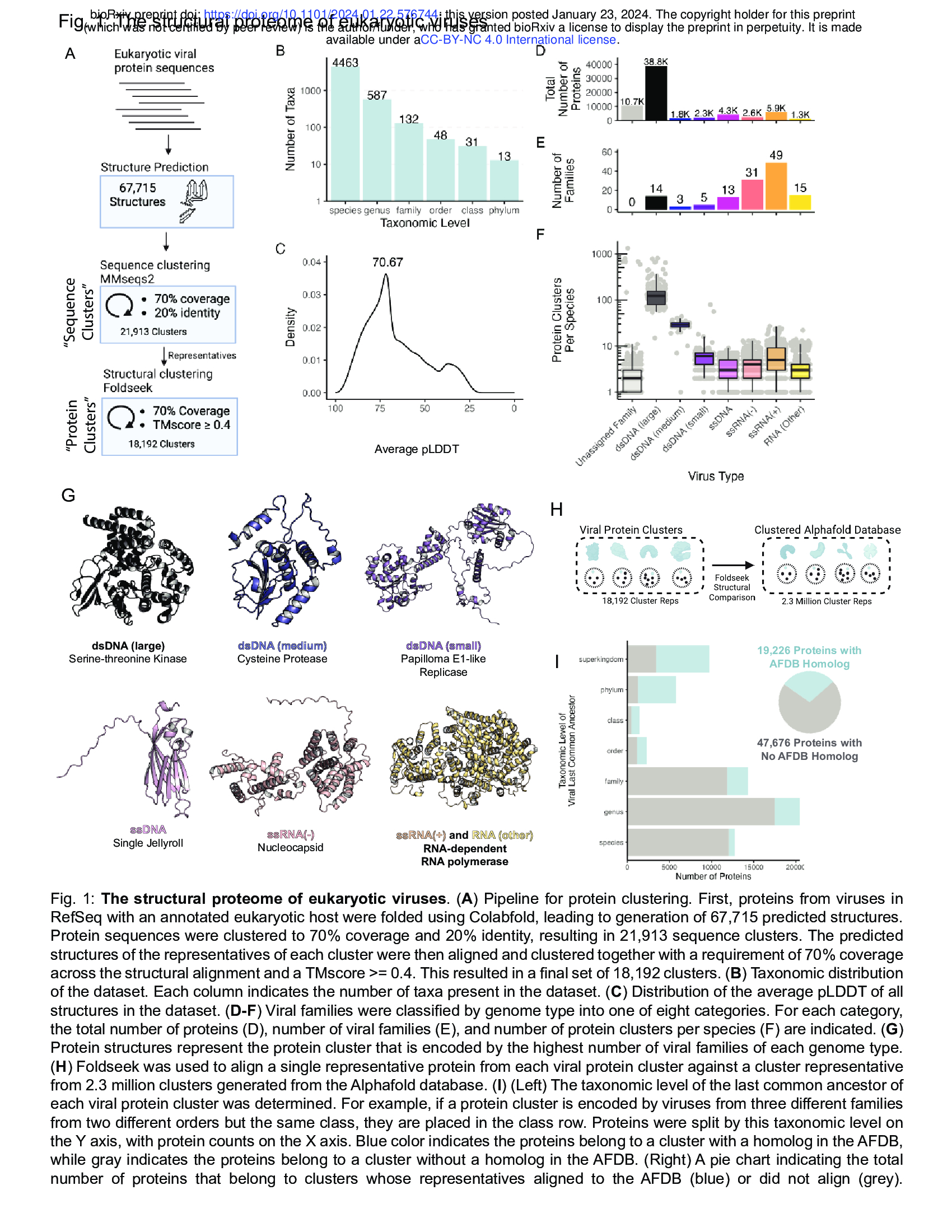
- The structural proteome of eukaryotic viruses /ˈproʊ.ti.oʊm/ (Fig. 1 + Supplementary Fig. 1)
[ColabFold is advanced version of AlphaFold (https://steineggerlab.com/en/)]
[Viral protein seqeunces --> structure prediction --> Seqeunce clustering using MMseqs2 --> Structural clustering using Foldseek]
- Eukaryotic viral protein sequences --> Structure prediction (67715 Strucutures) using Colabfold --> Sequence clustering using MMseqs2 (70% Coverage, 20% identity) --> Structural clustering using Foldseek (70% Coverage and TMscore>=0.4)
- This dataset includes a large diversity of viruses, including 4,463 species from 132 different viral families (Fig. 1B, C).
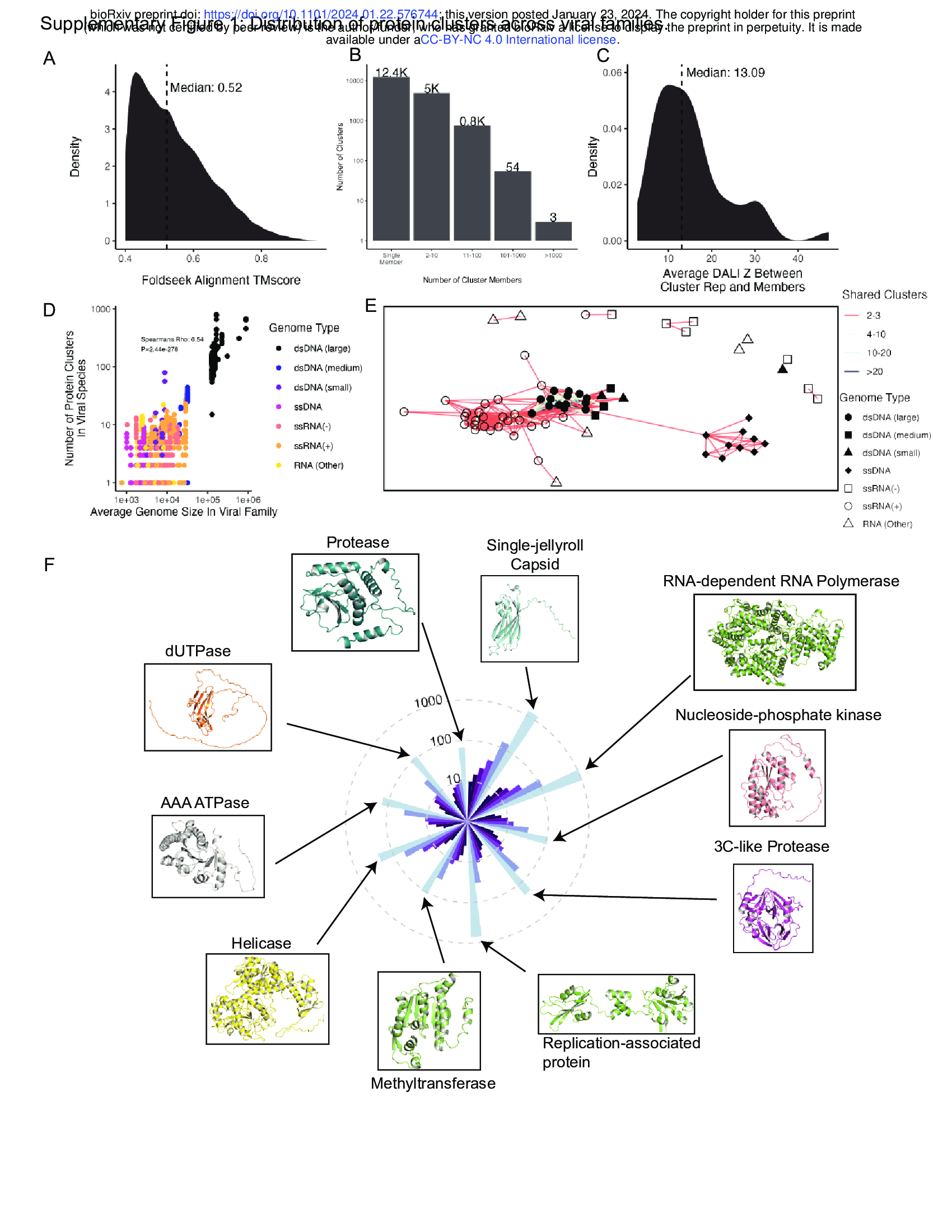
- Clusters are structurally consistent, as implementing DALI 24 to align cluster representatives to each member for clusters with at least 100 members yields a median cluster-average DALI Z score of 13.1 (Supplementary Fig. 1C). DALI Z-scores above 8 indicate
two proteins are likely homologous 25.
- We investigated how well this database represents viral diversity, and if it reconstitutes core viral hallmark genes.
- We grouped viral families into viral genome types based on their Baltimore classes26 with slight modifications – DNA viruses were split into large, medium, and small groupings based on their average genome length, while RNA viruses without single-stranded positive- or negative- sense genomes were grouped into the RNA (Other) category.
- Large double-stranded DNA (dsDNA) viruses have the most protein clusters per species and, despite constituting only 14 of the 132 viral families in the dataset, account for the majority of viral proteins (Fig. 1D, F).
- As expected, protein cluster count correlates strongly with genome size (Supplementary Fig. 1D).
- With their larger genomes, dsDNA viruses have the capacity to encode more auxiliary genes without sacrificing genome stability.
- RNA viruses make up a large fraction of the families present in the dataset, but a smaller fraction of the total proteins (Fig. 1E, F).
- Structural homology between viral families with a similar genome type is common, with large dsDNA viruses sharing many protein folds (Supplementary Fig. 1E).
- As expected, the predominant protein clusters in the dataset as a whole (Supplementary Fig. 1F) and within each genome type (Fig. 1G) are largely involved in fundamental aspects of the viral life cycle.
- These include the single jellyroll fold, which comprises viral capsids ['kæpsid] and is present in viruses of all genome types.
- The double jellyroll fold also comprises viral capsids, although it is restricted to dsDNA viruses27.
- RNA viral families often encode nucleocapsids, responsible for packaging of viral RNA, and RNA-dependent RNA polymerases (RdRPs) responsible for genome replication.
- While the RdRP is universally conserved in RNA viruses, it is split amongst multiple protein clusters due to variation in protein length.
- In contrast, small dsDNA viruses such as papillomaviruses and polyomaviruses encode a viral replicase with conserved origin-binding and helicase domains.
-! Altogether, we find that our structural database successfully reconstitutes conserved viral proteins across diverse viral subtypes.
- The pLDDT (predicted Local Distance Difference Test) value is a metric used in protein structure prediction, particularly by tools like AlphaFold, to assess the confidence in the predicted positions of amino acids in a protein structure. pLDDT scores range from 0 to 100, where higher scores indicate higher confidence in the accuracy of the predicted structure at specific positions. Scores above 90 are considered very high confidence, scores between 70 to 90 indicate confidence, scores between 50 to 70 are low confidence, and scores below 50 are very low confidence, suggesting that the model is uncertain about those regions of the protein structure.
- Viral Protein Clusters ---Foldseek structural comparison---> Clustered Alphafold Database (human proteins)
#viral protein cluster
- We next investigated the taxonomic distribution of viral protein clusters.
- We conducted structural alignments of viral protein cluster representatives against 2.3 million cluster representatives from the entire Alphafold Database (AFDB)3 (Fig. 1H).
- For each virus protein cluster, we determined the last common ancestor of viruses encoding a cluster member.
- We found that 29% of protein clusters are present in multiple viral families, the majority of which are present in the Alphafold Database, suggesting that they are evolutionarily ancient (Fig. 1I).
- In addition, we found that 62% of viral proteins (or 55% of proteins from non-singleton clusters) are restricted to a single viral family and lack homologs in the AFDB (Fig. 1I). This shows that viral evolution generates substantial numbers of novel proteins that are absent from current structure
databases.
2.Structural similarities between annotated and uncharacterized viral proteins (Fig. 2+Supplementary Fig. 2)
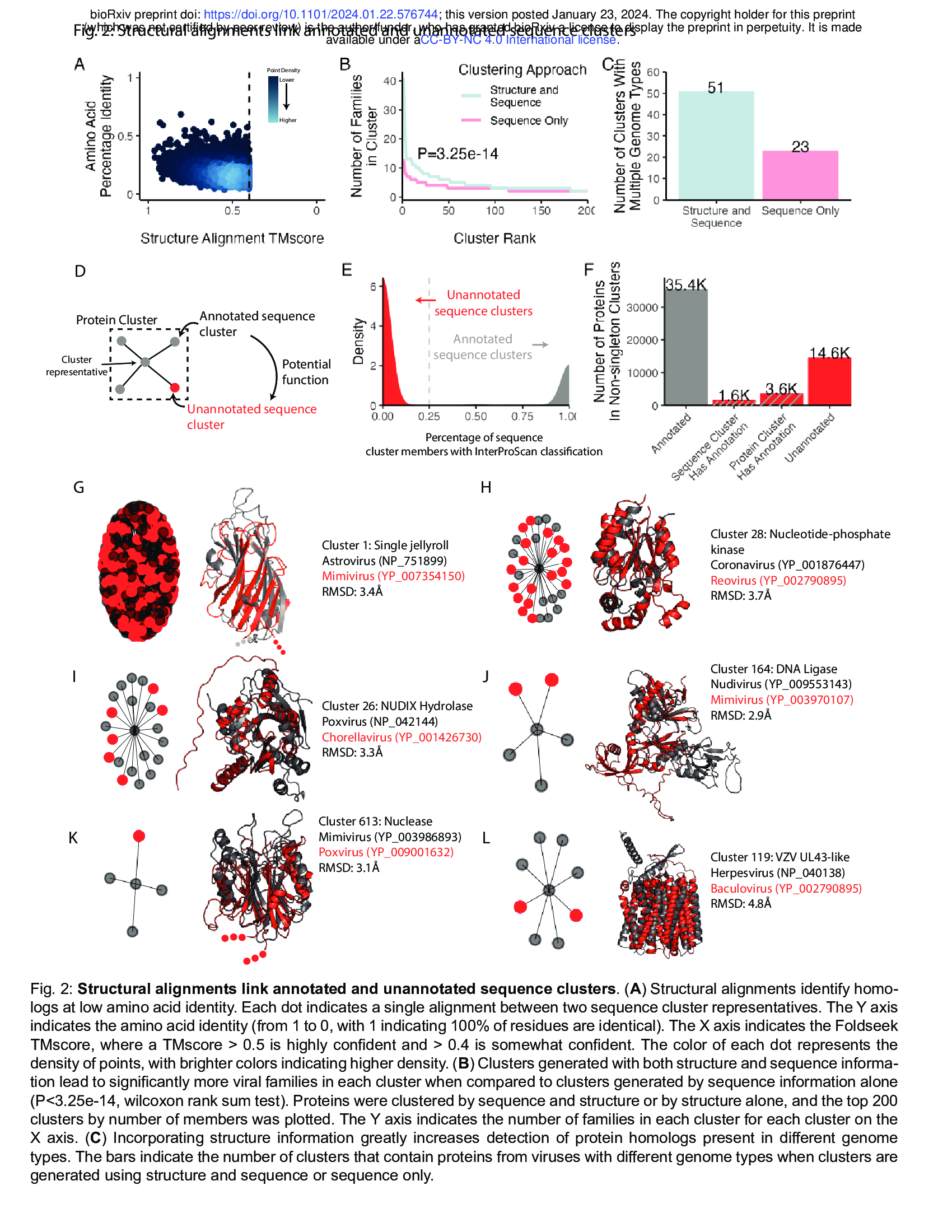
- We investigated the ability of structural alignments to identify relationships not apparent from protein sequence alone.
- We found that many representatives of sequence clusters are structurally similar despite low sequence similarity (Fig. 2A). --> the structure is more conserved than seqeunce level?
- In fact, adding structural information to protein clustering efforts leads to more taxonomically diverse protein clusters, with significantly more viral families per cluster (Fig. 2B). : with the structure and seqeunce, we have much more viral families per cluster!
- This is especially important for finding homology between proteins from divergent viruses, resulting in a substantial increase in protein clusters encompassing proteins encoded by viruses of different genome types (Fig. 2C). 51 vs 23
- We asked if [structural alignments] can link poorly-annotated sequence clusters with those that are more annotated (Fig. 2D). We used the sequence-based classifier InterProScan28 to assign all proteins Pfam29, CDD30, and TIGRFAM31 classifications.
- Sequence clusters contain almost entirely annotated or entirely unannotated members, resulting in a bimodal distribution of sequence clusters (Fig. 2E).
- Of the proteins in clusters with more than one member, over 25% ( 5.2/(1.6+3.6+14.6)=0.26 ) of unannotated proteins are located in either an annotated sequence cluster or a protein cluster that contains an annotated sequence cluster (Fig. 2F).
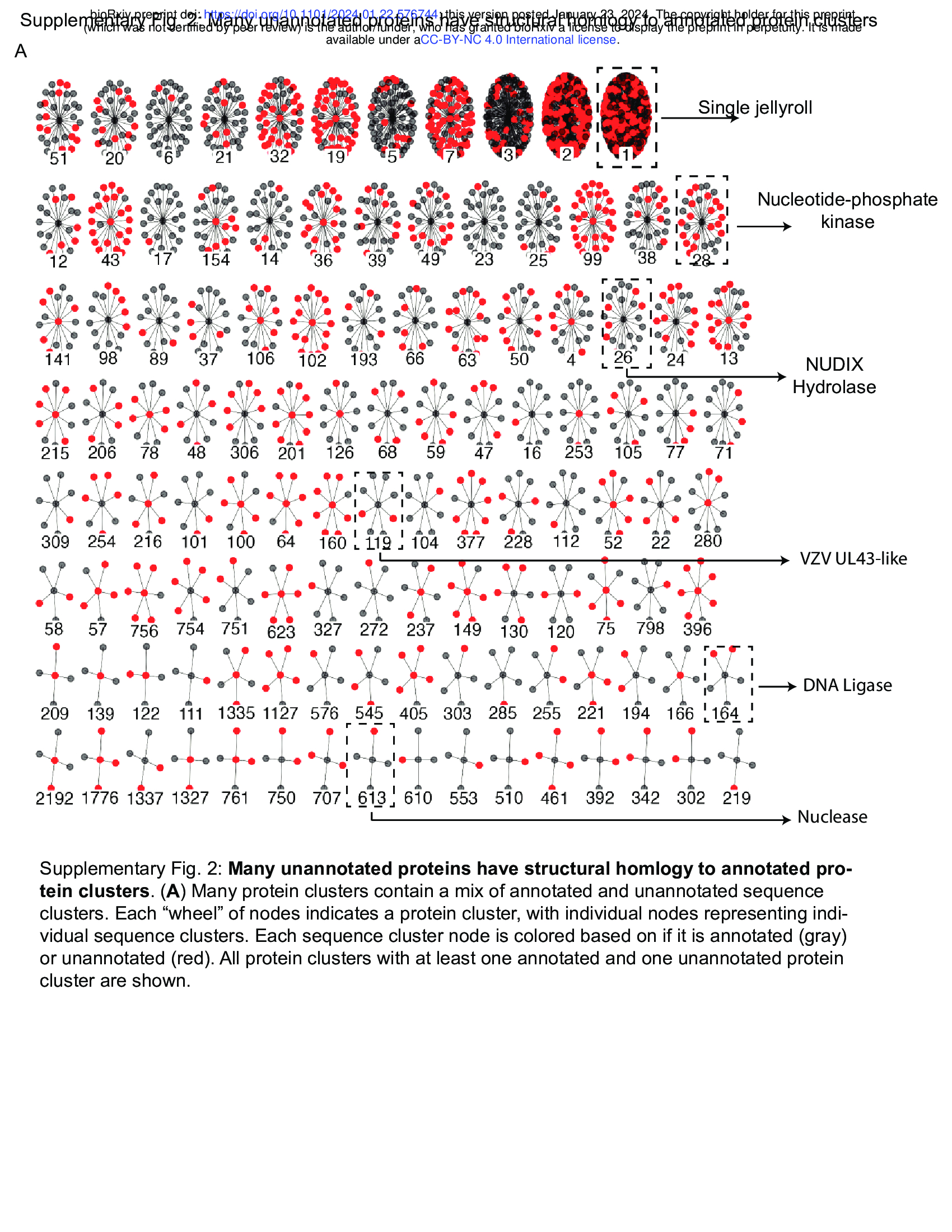
- Many protein clusters encompass a mixture of annotated and unannotated sequence clusters (Supplementary Fig. 2). Supplementary Fig. 2: Many unannotated proteins have structural homlogy to annotated protein clusters.
- We find that these connections between sequence clusters are useful to determine putative functions of poorly characterized proteins across the virome. For example, while the single jellyroll fold is the most abundant protein cluster, many members of this cluster are not correctly annotated (Fig. 2G). Many other protein clusters include both annotated and unannotated sequence clusters, including clusters encoding enzymes such as nucleotide-phosphate kinases (Fig. 2H), NUDIX Hydrolases (Fig. 2I), DNA ligases (Fig. 2J), and nucleases (Fig. 2K).
- One cluster of note includes members that resemble the UL43 family of late herpesvirus proteins (Fig. 2L), which will be discussed later.
- Together, these results demonstrate that large scale clustering based on sequence plus predicted structure enables functional inference of poorly characterized viral proteins.
#The Dali database is a structural classification based on precomputed all-against-all structural similarities within the PDB.
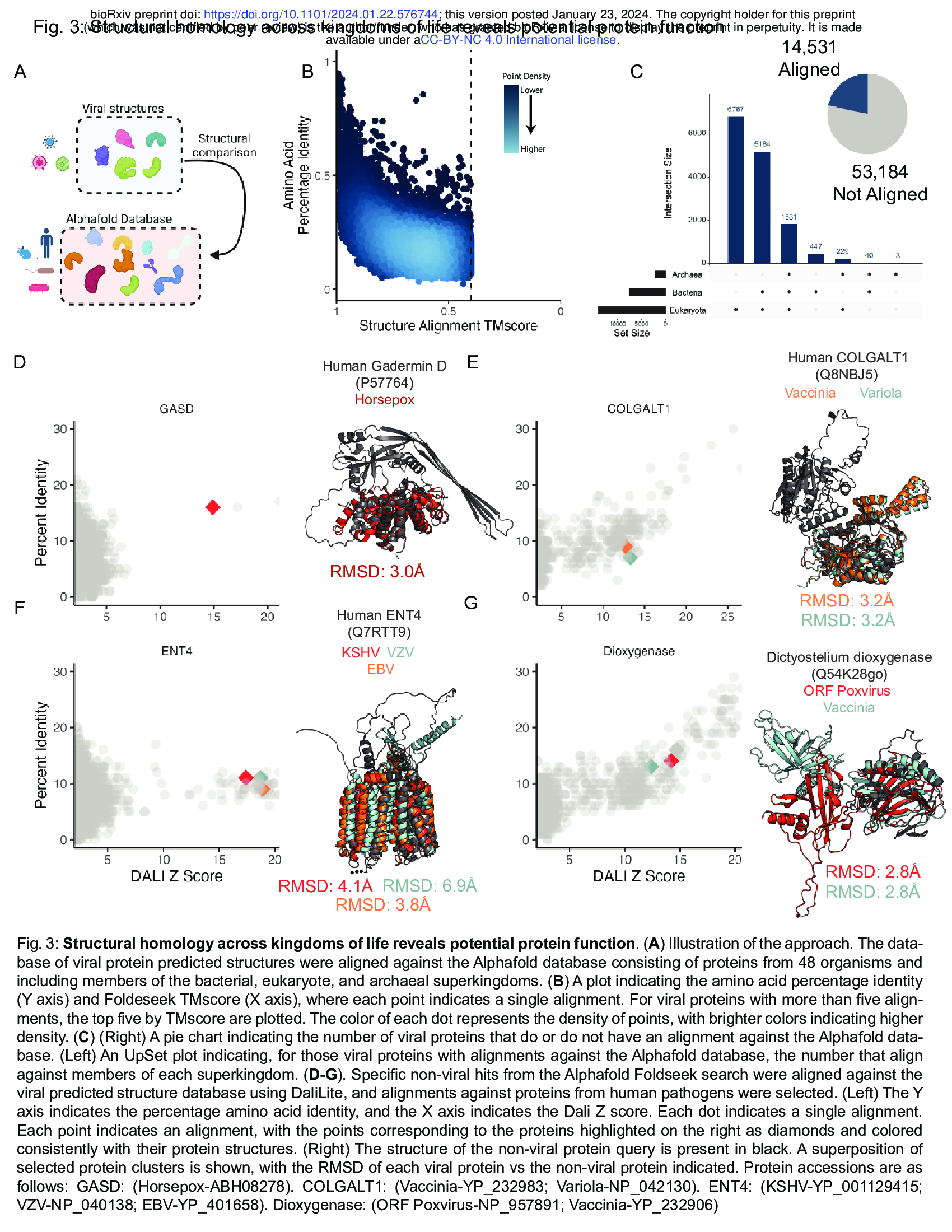
- Structural alignments suggest functions of human pathogen proteins ((Fig. 3)comparisions of viral (our databases) and non-viral proteins (Alphafold Databases) using Foldseek!)
Foldseek faster than TM-align and Dali!!!!!
#- Foldseek is a tool designed for fast and sensitive comparison of large sets of protein structures. It uses a novel approach by encoding structures as sequences over a 20-state 3D interaction alphabet, enabling comparisons through sequence alignments. This method significantly speeds up structural comparisons, making Foldseek much faster than traditional structural aligners like TM-align and Dali, while maintaining high sensitivity. Foldseek is available as open-source software and also through a webserver, facilitating rapid and accurate protein structure searches
#- Using Dali for structural comparison of proteins
- Unlike nucleotide or protein sequence, structural features are often conserved over large evolutionary timescales.
- Thus, we investigated if alignment between predicted viral and non-viral protein structures can offer insight into the function of poorly-annotated proteins encoded by human pathogens.
- To do this, we used Foldseek (for comparisisons) to align [our virus protein structure database!!!!] with the initial release of the Alphafold Database, which contains over 300,000 proteins from 21 organisms across eukaryotes, bacteria, and archaea2 (Fig. 3A).
- This revealed pervasive structural homology between viral and non-viral proteins, with high structural similarity in the face of low amino acid identity (Fig. 3B).
- Ultimately, 14,531 predicted viral proteins have an alignment to a member of the Alphafold database, with the majority of alignments being against proteins encoded by eukaryotes (Fig. 3C).
- These alignments include proteins that are unannotated but are encoded by human pathogens.
- To reduce rates of false negatives, we conducted a series of alignments using DALI24, which is slower than Foldseek but substantially more sensitive.
- First, we found that a set of proteins encoded by poxviruses are structurally similar to the auto-inhibitory domain of mammalian gasdermins (Fig. 3D)32.
-* In the context of "auto-inhibitory domain of mammalian gasdermins," the term "domain" refers to a specific part of a protein that has a distinct structure and function. In proteins like gasdermins, which are involved in cell death pathways, an auto-inhibitory domain acts as a regulatory segment. This domain can prevent the protein from acting until certain conditions are met, effectively inhibiting the protein's activity to regulate processes such as inflammation and cell death until it is appropriate for the protein to become active.
- Similarly, several poxvirus proteins are structurally homologous to the human galactosyltransferase COLGALT1, thought to enable virus binding to
surface glycosaminoglycans during viral entry (Fig. 3E)33.
- Next, we found that human herpesviruses proteins, including the protein BMRF2 from Epstein Barr herpesvirus (EBV) and Varicella zoster virus (VZV), share structural similarity with the human equilibrative nucleoside transporter ENT4 (Fig. 3F).
- EBV conducts substantial remodeling of host cell metabolism during viral infection34, and this finding suggests a potential metabolic role in addition to BMRF2 involvement in viral attachment35.
- In addition, transport of antiviral nucleoside analogues such as valacyclovir are mediated by nucleoside transporters36,37, raising questions about the
interplay between this protein and valacyclovir during VZV infection. These proteins belong to a cluster of proteins similar to the UL43 family of late herpesvirus proteins, some of which are unannotated (Fig. 2J).
- In addition, we observed structural homology of Poxvirus C4-like proteins with eukaryotic dioxygenases (Fig. 3G). Vaccinia virus C4 is notable for antagonizing several innate immune pathways. C4 directly binds the pattern recognition receptor DNA-PK, blocking DNA binding and immune signaling through that pathway38. In addition, C4 inhibits NF-κB signaling downstream at or downstream of the IKK complex, but the mechanism of this inhibition is unknown39. Future studies are required to determine if its dioxygenase-like fold is involved in its innate immune antagonism.
- Altogether, these findings illustrate the ubiquity of structural homology between viral and non-viral proteins and show that this homology can be used to predict potential functions of poorly characterized viral proteins.
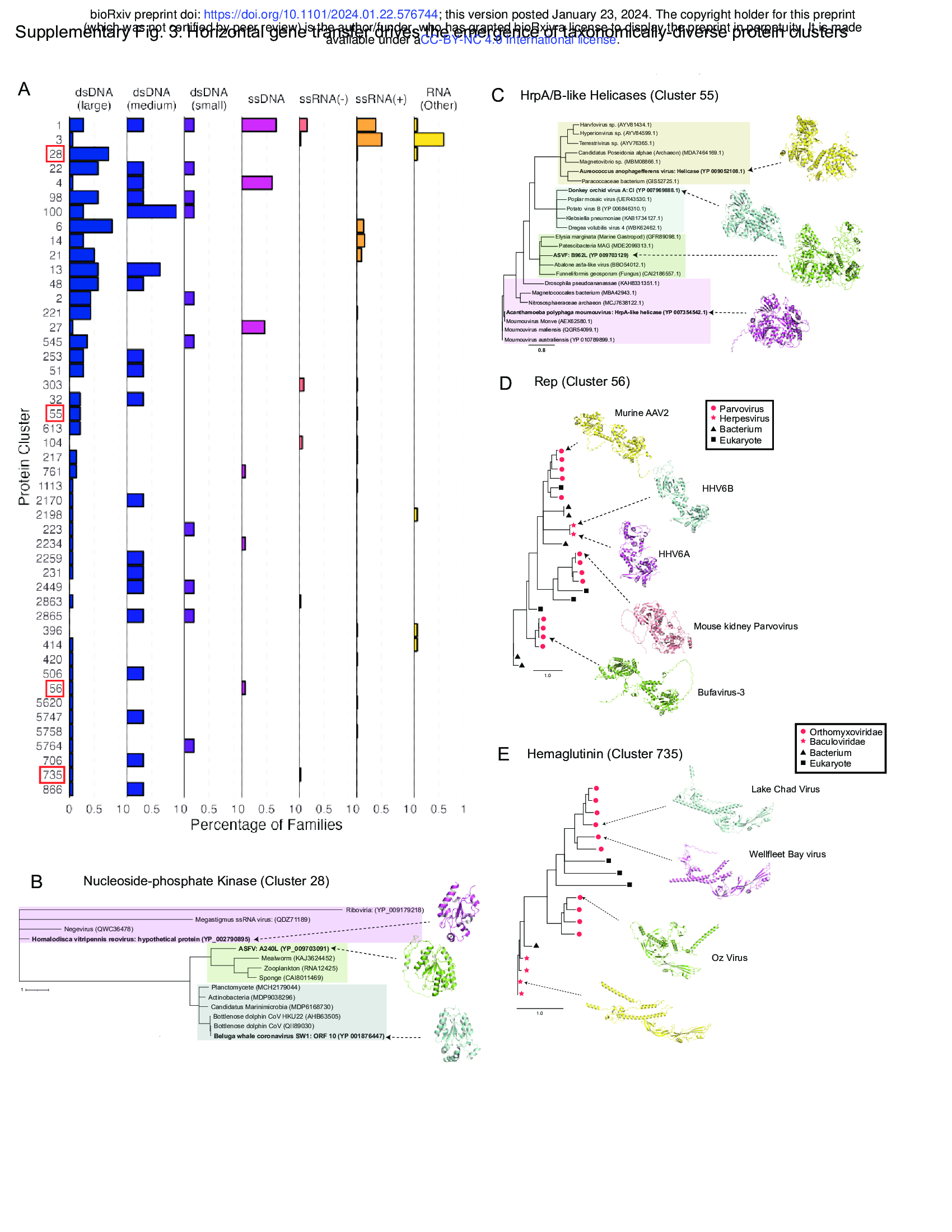
- Horizontal gene transfer creates taxonomically-diverse protein clusters (Supplementary Fig. 3)
-* What does "domain" in protein structure mean? While alpha-helices and beta-sheets are elements of secondary structure within a protein, a domain is a higher order of structure that often consists of multiple secondary structure elements arranged in a specific configuration. Domains can serve various functions, such as catalytic activity, binding to other molecules, or regulatory roles, and a single protein can contain multiple domains each performing different functions.
- While we found that some protein clusters contain members encoded by viruses of different genome types, the evolutionary origin of such conservation is unclear.
- Many of these protein clusters are predominantly encoded by viruses of a single genome type but expressed in a small minority of viruses of a different genome type (Supplementary Fig. 3A).
- This observation is consistent with virus-virus or host-virus horizontal gene transfer.
- To explore this possibility, we conducted Blast40 searches of sequence cluster representatives against viral- and non-viral protein databases and constructed phylogenetic trees of the top hits.
- We found that nucleoside-phosphate kinases in cluster 28 show a polyphyletic distribution with homologs in different viruses showing amino acid similarity to distinct sets of non-viral proteins (Supplementary Fig. 3B). Homalodisca vitripennis reovirus
- There is a similar pattern with HrpA/B-like helicases in Cluster 55, with helicases in different viral families showing amino acid similarity to distinct sets of non-viral organisms (Supplementary Fig. 3C).
- These patterns are consistent with horizontal gene transfer from non-viral hosts.
- In contrast, other taxonomically distributed protein clusters such as cluster 56 (encoding parvovirus Rep proteins with homologs in some human herpesviruses) and cluster 735 (encoding a hemagglutinin lineage present in baculoviruses and some orthomyxoviruses) display a monophyletic taxonomic distribution consistent with horizontal gene transfer between viruses (Supplementary Fig. 3D, E).
- These data suggest that many protein clusters that contain proteins from viruses of different genome types arise from horizontal gene transfer, both from viral and non-viral sources.
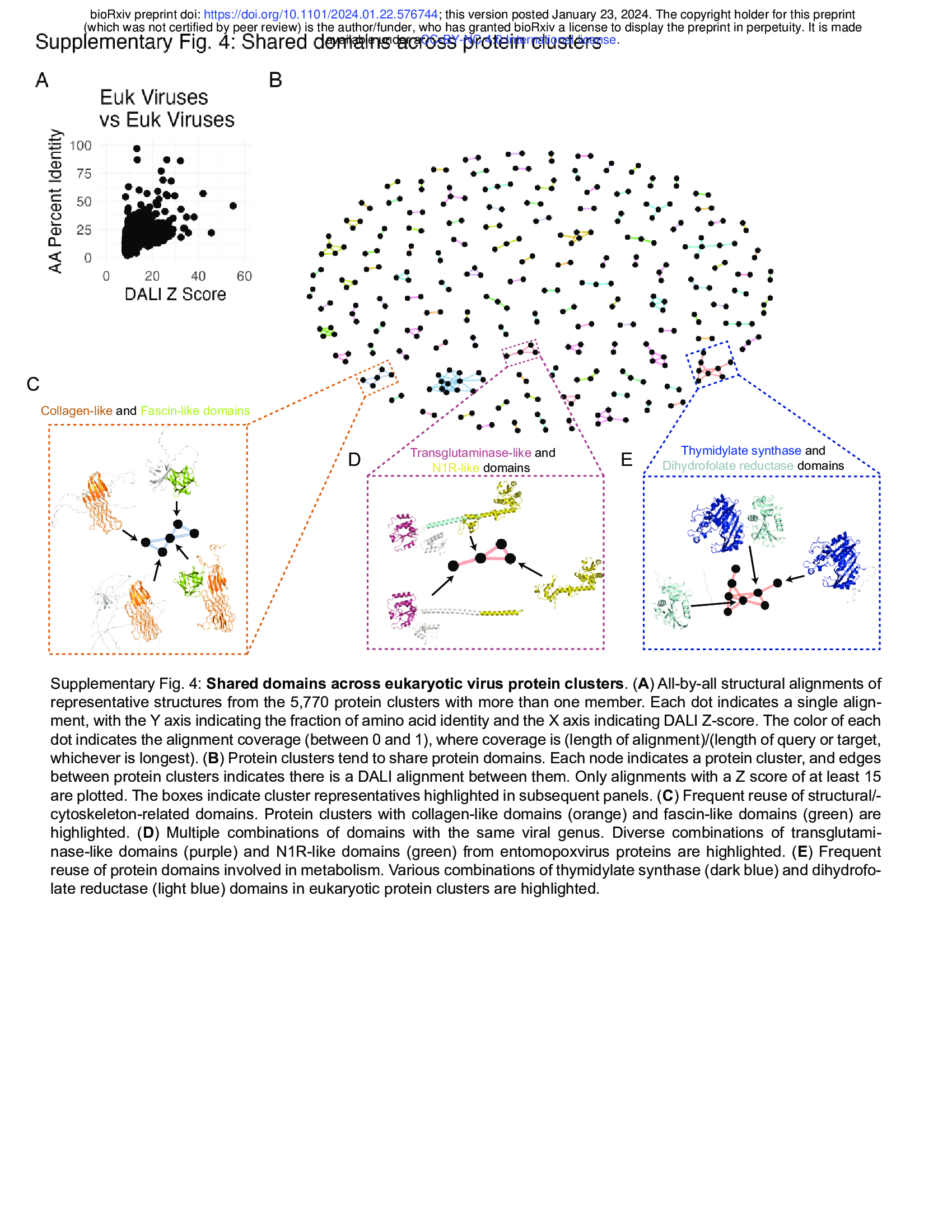
- Structural alignments identify shared functional domains (Supplementary Fig. 4)
- We constructed protein clusters with a strict 70% coverage requirement, leaving open the possibility that individual domains can be identified through structure comparison3.
- We reasoned that protein domains present within multiple protein clusters may have particular biological importance.
- We used DALI to conduct all-by-all alignments of the representatives of all protein clusters having more than one member.
- This revealed substantial protein similarity with many alignments having Z scores greater than 8, indicating high confidence of structural homology25
(Supplementary Fig. 4A).
- Protein clusters ultimately fall into a network of shared domains (Supplementary Fig. 4B).
- Here, distinct domains are often shared across protein clusters in context with various combinations of other domains, which can be seen with domains involved in interaction with the cytoskeleton (Supplementary Fig. 4C) and in metabolism (Supplementary Fig. 4D,E) in eukaryotic viruses and phage.
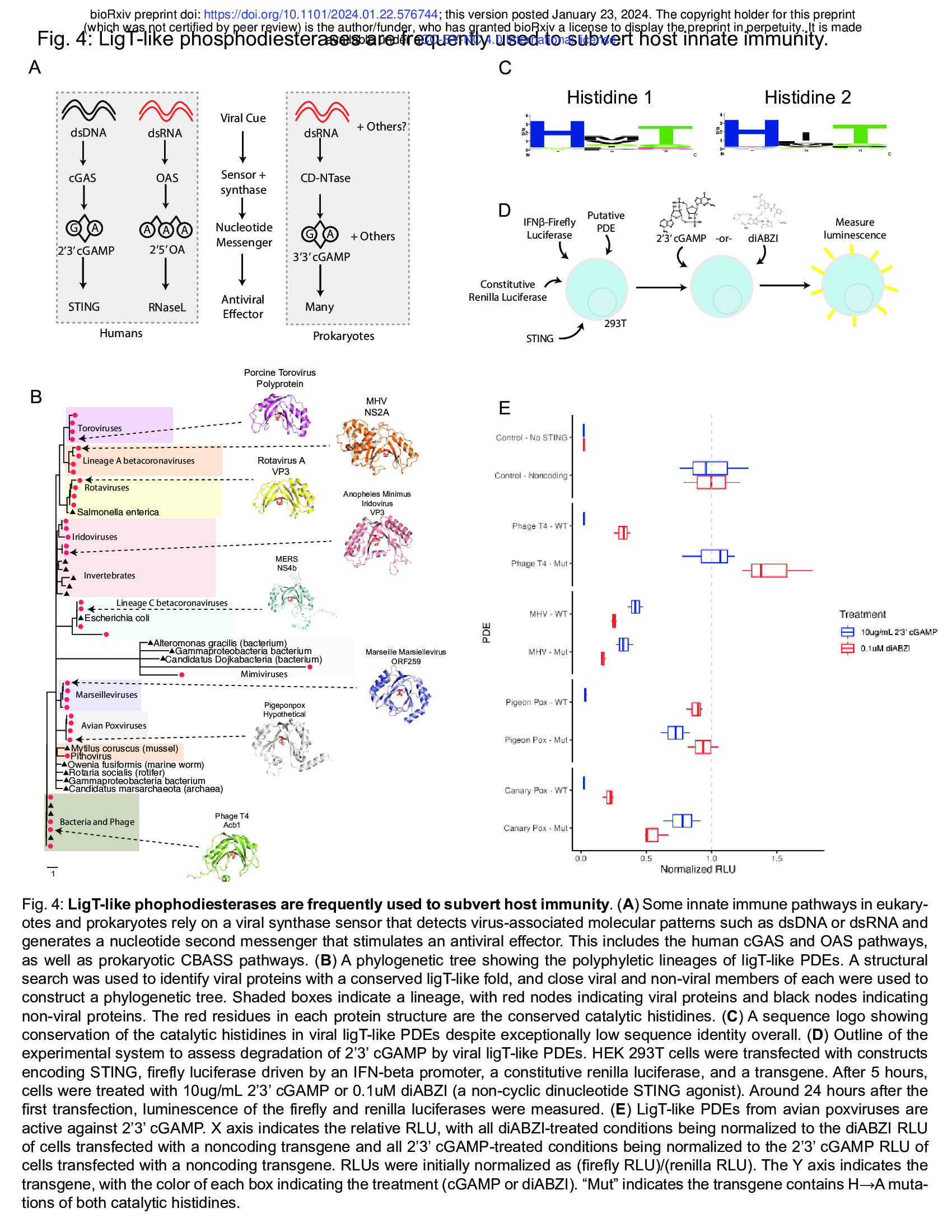
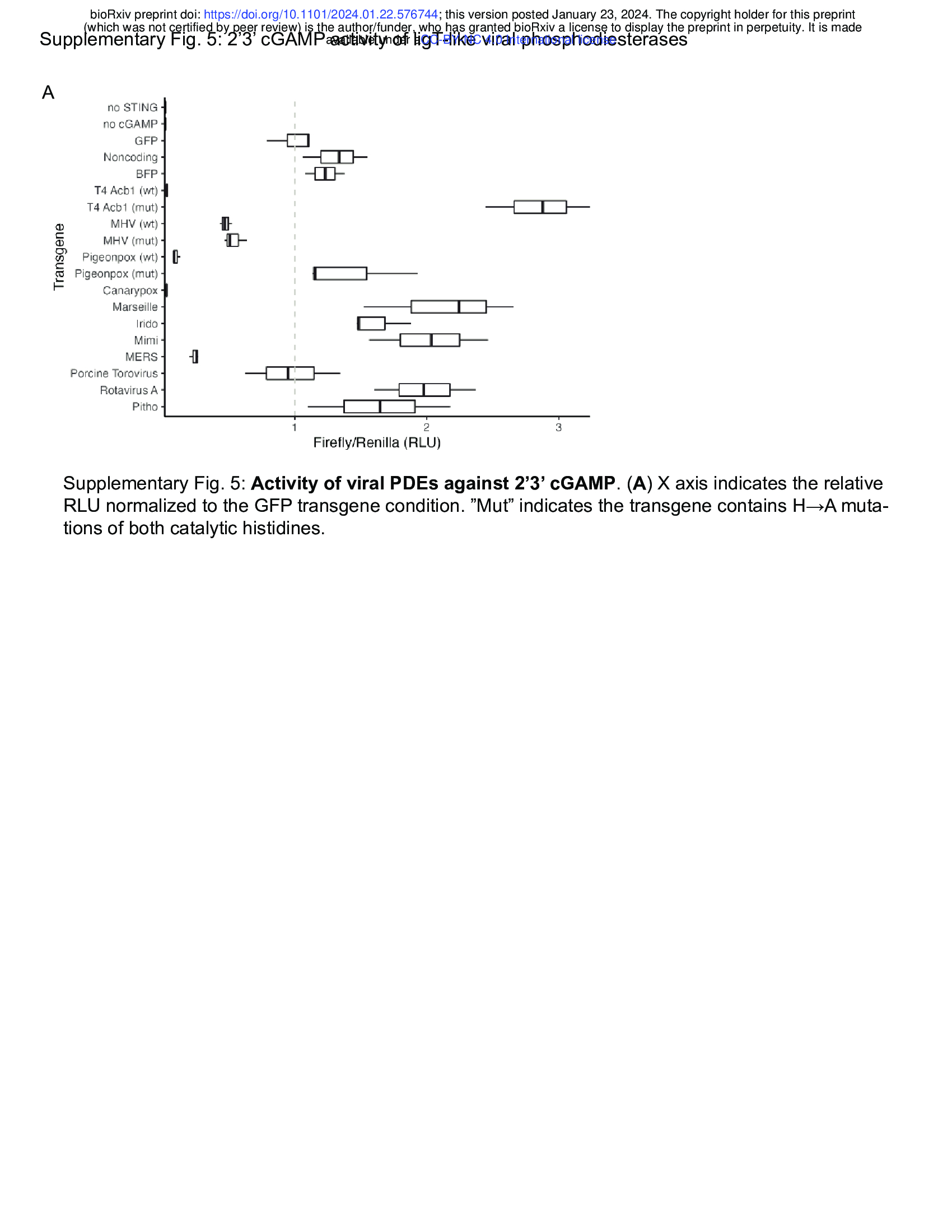
- Structural homology reveals phosphodiesterases that degrade 2’3’ cGAMP (e.g. LigT-like phophodiesterases (Fig. 4 + Supplementary Fig. 5))
Many aspects of eukaryotic and prokaryotic immunity have a shared origin41. One set of related
pathways are the mammalian cGAS-STING and OAS pathways and prokaryotic
Cyclic-oligonucleotide-based anti-phage signaling systems (CBASS). In both cases, a protein
sensor detects a viral cue and generates a nucleotide second messenger, which activates a
downstream antiviral effector (Fig. 4A). In the case of the cGAS (cyclic GMP-AMP synthase)
pathway, cGAS recognizes cytoplasmic double-stranded DNA and generates 2’3’ cyclic
GMP-AMP (2’3’ cGAMP). Many cGAS/DncV-like nucleotidyltransferases (CD-NTases) in
prokaryotic CBASS’ make a similar second messenger, 3’3’ cGAMP, in response to viral cues42.
In contrast, OAS (oligoadenylate synthase) recognizes double-stranded RNA and generates
linear 2’5’ oligoadenylates (2’5’ OA)43,44. In prokaryotes, phage T4 encodes the ligT-like PDE
anti-CBASS protein 1 (Acb1), which degrades 3’3’ cGAMP and a variety of other cyclic
nucleotide substrates including 2’3’ cGAMP45.
In eukaryotes, several RNA viruses encode PDEs that degrade 2’5’ OA46. Interestingly,
we find that these PDEs have a ligT-like fold similar to Acb1. Given the conserved use of
ligT-like PDEs in viral anti-immunity, we investigated their distribution and phylogeny. Structural
searches revealed many different branches of ligT-like PDEs are present in eukaryotic viruses
(Fig. 4B). Notably, there are multiple independent branches of ligT-like PDEs in RNA viruses.
Linage A betacoronaviruses and Toroviruses share a clade of PDEs that is similar to the PDEs
present in Rotaviruses. Surprisingly, lineage C betacoronaviruses contain a distinct branch of
PDEs (Fig. 4A)47. This suggests that there were two independent PDE acquisition events within
the betacoronavirus genus, showing the strong selective pressure for betacoronaviruses to
evade the OAS pathway. We find that some large DNA viruses also contain ligT-like PDEs.
Despite the extreme amino acid variability across the ligT-like PDE tree there is near-universal
conservation of the two catalytic histidines (Fig. 4C), with the exception of the Mimivirus ligT-like
branch.
The presence of ligT-like PDEs in large DNA viruses raises the question of whether they
have an anti-immune function. While the RNA-sensing OAS pathway is commonly targeted by
ligT-like PDEs of RNA viruses, there is likely less pressure for large DNA viruses to target OAS.
Thus, we tested whether ligT-like PDEs encoded by large DNA viruses have activity against 2’3’
cGAMP. To address this question, we generated a synthetic STING circuit in 293T cells48,49 (Fig.
4D). Here, STING can be activated by treatment with 2’3’ cGAMP or the non-nucleotide STING
agonist diABZI50, which will lead to expression of firefly luciferase in a STING-dependent
manner. We expect that a viral PDE that targets 2’3’ cGAMP should be able to inhibit 2’3’
cGAMP- but not diABZI-mediated STING activity. Testing representative PDEs from each
branch revealed that while PDEs encoded by RNA viruses and other large DNA viruses have
only limited activity against 2’3’ cGAMP, PDEs encoded by avian poxviruses have very potent
activity against 2’3’ cGAMP (Supplementary Fig. 5A). We found that the ligT-like PDEs encoded
by Pigeonpox and Canarypox very potently restrict STING signaling stimulated by 2’3’ cGAMP
but have limited activity against diABZI-mediated STING signaling (Fig. 4E). Furthermore,
mutation of the catalytic histidines substantially reduces activity (Fig. 4E). Avian poxvirues are
notable for their lack of Poxin51, the other 2’3’ cGAMP phosphodiesterase encoded by
poxviruses, showing the strong selective pressure for poxviruses to evade cGAS-STING
immunity. Altogether, we have leveraged structure homology to discover a novel mechanism of
2’3’ cGAMP degradation by eukaryotic viruses and find that cGAMP targeting by ligT-like PDEs
is a pan-viral mechanism of anti-immunity.
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Kraken2 Installation and Usage Guide
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Should the inputs for GSVA be normalized or raw?
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- Risks of Rebooting into Rescue Mode
- 足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
基因组与表型特性研究:一株药物敏感的鲍曼不动杆菌显示出增强的毒力相关特性和应激耐受性