Easyfig usage
gene_x 0 like s 958 view s
Tags: software
-
How to use the program?
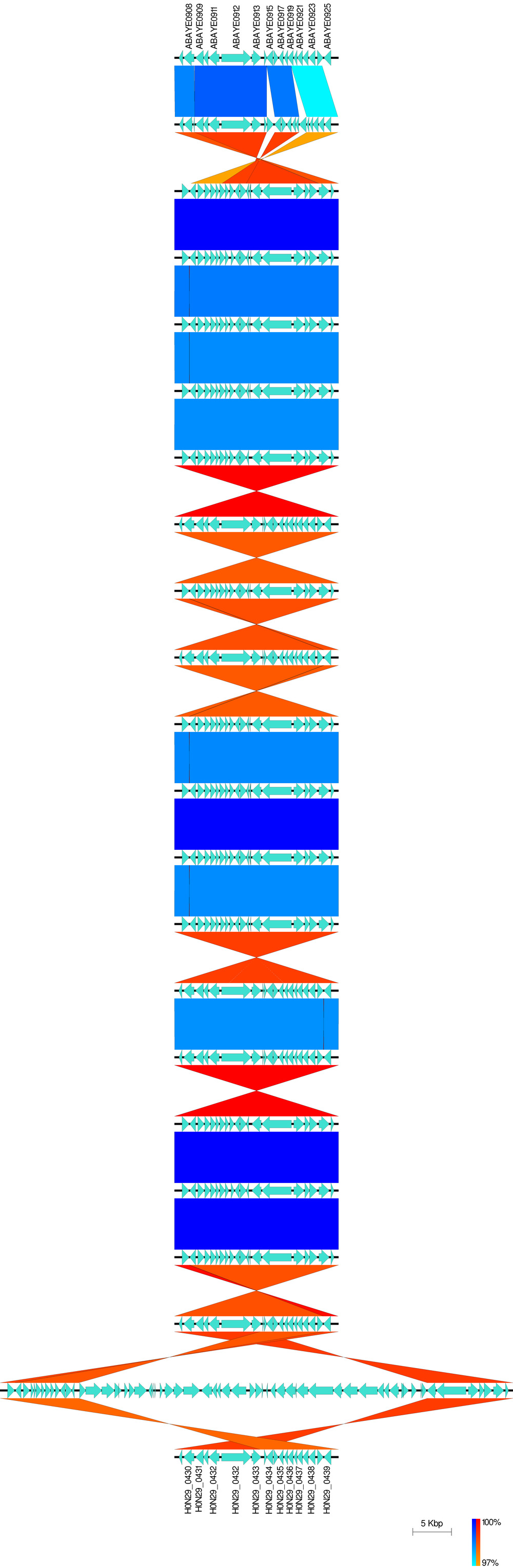
D: Zoom in on a ~15kb subregion at one end of the sequences Access the subregion window from the Image dropdown menu; click on file 01 and then enter 32599 and 50099 in the Min and Max "Range" boxes located directly under the list of annotation files Click (change cutoffs) Click on file 02 and then enter 1 and 15000 in the Min and Max "Range" boxes and click (change cutoffs) IMPORTANT: remember that the second Annotation file (LT2_Gifsy.gbk) has been inverted, This has been taken into account when entering the subregion range Close the subregion window with (close) Click on ( Generate blastn Files ) to generate the BLAST comparison files for these subregions (a pop-up box will ask you where you want to save the BLAST files – default is usually in the Easyfig_example1_files folder) CREATE FIGURE as described in part 1A. In the new image, the scale is still 5000 bp, but the image is zoomed in on the variable region. The minimum BLAST identity value is shown in the yellow dialog box after each figure is drawn – this can be used to calibrate the BLAST identity scale shown on the right (i.e. in this case the matches range from 100% (darkest) to 85% (lightest)). As described in the manual there are many ways to customise the image further e.g. the following list shows a few of the options available: * Feature rendering type from arrows to boxes or pointers * Colours of any of the features * Thickness of any lines * Height of BLAST matches * Height of features * Width of figure * Type of image file (bmp is default, but svg [scalable vector graphics] files can be produced by changing the file type) * Add a gene legend or label the genes** #https://github.com/gamcil/clinker - clinker to finish the last part of Data_PaulBongarts_S.epidermidis_HDRNA: namely compare gene order in the a part of genbank ( * genomic rearrangements (e.g. SCCmec deletions, ACME deletions, agr insertions) #Staphylococcal Cassette Chromosome mec #arginine catabolic mobile element (ACME) #Prevalence and genetic diversity of arginine catabolic mobile element (ACME) in clinical isolates of coagulase-negative staphylococci: identification of ACME type I variants in Staphylococcus epidermidis. Fig. 1. A schematic drawing of genetic structures of ACME (a region from the arc to opp3 cluster, or corresponding genetic components) among the three DI subtypes (DI.1, DI.2, and DI.3: strains CNS266, CNS115, and CNS149, respectively), type I (strain USA300-FPR3757, accession number CP000255), type II (strain ATCC12228, accession number AE015929) and type DII (strain M08/0126, accession number FR753166). Putative ORFs of genes are represented by arrows colored with green (arc cluster), red (opp3 cluster), blue (a region between the arc and opp3 clusters in ACME I), or dark blue (genes in ACME II). The regions in light pink including the arc cluster indicate genetically identical areas to both ATCC12228 and USA300-FPR3757. The regions with light blue are identical to only ATCC12228, while those with light orange to USA300-FPR3757. White space regions between argR and SAUSA300_0072 show no sequence homology either to ATCC12228 or to USA300_FPR3757; however, these regions show 91–99% nucleotide identity among the three ACME subtypes. Regions colored with dark orange in the three ACME DI subtypes show=98% nucleotide sequence identity to each other. Regions colored with grey (type DI.1), purple or cyan (type DI.3) do not show high nucleotide identity (<98%) to cognate genes in other ACME types (Table S2.2). Positions of primers used for PCR profile (Tables 1 and 4) are shown with arrowheads under ACME I sequence. Collapse #https://mjsull.github.io/Easyfig/files.html #https://github.com/mjsull/Easyfig/wiki/ Easyfig # - Fig. 4. AdeIJK has fewer SNPs within it and lower levels of recombination surrounding it than AdeABC or AdeFGH. # - In total, 100 A. baumannii genomes were aligned against reference A. baumannii AYE (NC_010410.1) and the presence of polymorphisms and recombination was determined using Gubbins. # - (a,c,e) Magnified parts of the genome at each ade operon, showing the levels of SNPs (red and blue squares; red are ancestral SNPs) and recombination levels (the black line on the bottom; the higher the peak the more recombination). # - The right-hand panels (b, d and f) show the entire genome and the position of each different ade operon, which is highlighted in red beneath the label. #- All panels have an associated mid-point rooted phylogenetic tree created by Snippy to show the relatedness of the A. baumannii sequences. #- AdeIJK (a) has fewer SNPs and recombination than AdeABC (e) and AdeFGH (c), indicating it is highly conserved. -
Input Data
https://www.genome.jp/dbget-bin/www_bfind_sub?mode=bfind&max_hit=1000&locale=en&serv=kegg&dbkey=genome&keywords=Acinetobacter+baumannii&page=1 T00667 aby; Acinetobacter baumannii AYE --> GCA_000069245.1_ASM6924v1 T00660 abm; Acinetobacter baumannii SDF --> GCA_000069205.1_ASM6920v1 T00710 abc; Acinetobacter baumannii ACICU --> GCA_000018445.1_ASM1844v1 T00793 abn; Acinetobacter baumannii AB0057 --> GCA_000021245.2_ASM2124v2 T00795 abb; Acinetobacter baumannii AB307-0294 T01819 abx; Acinetobacter baumannii 1656-2 T01820 abz; Acinetobacter baumannii MDR-ZJ06 T01908 abd; Acinetobacter baumannii TCDC-AB0715 T02045 abr; Acinetobacter baumannii MDR-TJ T02261 abh; Acinetobacter baumannii TYTH-1 T02491 abad; Acinetobacter baumannii D1279779 T02726 abj; Acinetobacter baumannii BJAB07104 T02727 abab; Acinetobacter baumannii BJAB0715 T02728 abaj; Acinetobacter baumannii BJAB0868 T02954 abaz; Acinetobacter baumannii ZW85-1 T03374 abk; Acinetobacter baumannii AbH12O-A2 T03375 abau; Acinetobacter baumannii AB030 T03376 abaa; Acinetobacter baumannii AB031 T03377 abw; Acinetobacter baumannii AC29 T03519 abal; Acinetobacter baumannii LAC-4 T00486 acb; Acinetobacter baumannii ATCC 17978 --> GCA_000015425.1_ASM1542v1 -
After downloading the Genbank, perform the following commands to calculate the Subregions positions.
~/Scripts/genbank2fasta.py A.baumannii_AYE.gbk ~/Scripts/genbank2fasta.py A.baumannii_SDF.gbk ~/Scripts/genbank2fasta.py A.baumannii_ACICU.gbk ~/Scripts/genbank2fasta.py Acinetobacter_baumannii_AB0057.gbk ~/Scripts/genbank2fasta.py Acinetobacter_baumannii_AB307-0294.gbk ~/Scripts/genbank2fasta.py Acinetobacter_baumannii_1656-2.gbk ~/Scripts/genbank2fasta.py Acinetobacter_baumannii_MDR-ZJ06.gbk ~/Scripts/genbank2fasta.py Acinetobacter_baumannii_TCDC-AB0715.gbk ~/Scripts/genbank2fasta.py Acinetobacter_baumannii_MDR-TJ.gbk ~/Scripts/genbank2fasta.py Acinetobacter_baumannii_TYTH-1.gbk ~/Scripts/genbank2fasta.py Acinetobacter_baumannii_D1279779.gbk ~/Scripts/genbank2fasta.py Acinetobacter_baumannii_BJAB07104.gbk ~/Scripts/genbank2fasta.py Acinetobacter_baumannii_BJAB0715.gbk ~/Scripts/genbank2fasta.py Acinetobacter_baumannii_BJAB0868.gbk ~/Scripts/genbank2fasta.py Acinetobacter_baumannii_ZW85-1.gbk ~/Scripts/genbank2fasta.py Acinetobacter_baumannii_AbH12O-A2.gbk ~/Scripts/genbank2fasta.py Acinetobacter_baumannii_AB030.gbk ~/Scripts/genbank2fasta.py Acinetobacter_baumannii_AB031.gbk ~/Scripts/genbank2fasta.py Acinetobacter_baumannii_AC29.gbk ~/Scripts/genbank2fasta.py Acinetobacter_baumannii_LAC-4.gbk ~/Scripts/genbank2fasta.py A.baumannii_ATCC17978.gbk cat [the first 4 gbk files] Acinetobacter_baumannii_AB0057.gbk_converted.fna Acinetobacter_baumannii_AB307-0294.gbk_converted.fna Acinetobacter_baumannii_1656-2.gbk_converted.fna Acinetobacter_baumannii_MDR-ZJ06.gbk_converted.fna Acinetobacter_baumannii_TCDC-AB0715.gbk_converted.fna Acinetobacter_baumannii_MDR-TJ.gbk_converted.fna Acinetobacter_baumannii_TYTH-1.gbk_converted.fna Acinetobacter_baumannii_D1279779.gbk_converted.fna Acinetobacter_baumannii_BJAB07104.gbk_converted.fna Acinetobacter_baumannii_BJAB0715.gbk_converted.fna Acinetobacter_baumannii_BJAB0868.gbk_converted.fna Acinetobacter_baumannii_ZW85-1.gbk_converted.fna Acinetobacter_baumannii_AbH12O-A2.gbk_converted.fna Acinetobacter_baumannii_AB030.gbk_converted.fna Acinetobacter_baumannii_AB031.gbk_converted.fna Acinetobacter_baumannii_AC29.gbk_converted.fna Acinetobacter_baumannii_LAC-4.gbk_converted.fna > all_converted.fasta #for file in 1.easyfig.fa 2.easyfig.fa 3.easyfig.fa 4.easyfig.fa; do (cat "${file}"; echo) >> all.easyfig.fa; done makeblastdb -in all_converted_.fasta -dbtype nucl #-max_target_seqs 1 blastn -db all_converted_.fasta -query ABAYE_RS05070.fasta -num_threads 15 -outfmt 6 -strand both -evalue 0.1 > ABAYE_RS05070_positions2.easyfig.out CU459141.1_Acinetobacter_baumannii_AYE 974325 975530 --> 964325 985530 CU468230.2_Acinetobacter_baumannii_SDF 854002 855207 --> 844002 865207 CP000863.1_Acinetobacter_baumannii_ACICU 2980675 2979470 --> 2969470 2990675 CP001921.1_Acinetobacter_baumannii_1656-2 3018236 3017031 --> 3007031 to 3028236 CP009257.1_Acinetobacter_baumannii_strain_AB030 1791300 1790095 --> 1780095 to 1801300 CP009256.1_Acinetobacter_baumannii_strain_AB031 3303170 3301965 --> 3291965 to 3313170 CP001182.2_Acinetobacter_baumannii_AB0057 3087319 3086114 --> 3076114 to 3097319 CP001172.2_Acinetobacter_baumannii_AB307-0294 974367 975572 --> 964367 to 985572 CP009534.1_Acinetobacter_baumannii_strain_AbH12O-A2 2890551 2889346 --> 2879346 to 2900551 CP007535.2_Acinetobacter_baumannii_strain_AC29 1283600 1284805 --> 1273600 to 1294805 CP003847.1_Acinetobacter_baumannii_BJAB0715 3060955 3059750 --> 3049750 to 3070955 CP003849.1_Acinetobacter_baumannii_BJAB0868 2950975 2949770 --> 2939770 to 2960975 CP003846.1_Acinetobacter_baumannii_BJAB07104 3043603 3042398 --> 3032398 to 3053603 CP003967.2_Acinetobacter_baumannii_D1279779 2755134 2753929 --> 2743929 to 2765134 CP007712.1_Acinetobacter_baumannii_LAC-4 976323 977528 --> 966323 to 987528 CP003500.1_Acinetobacter_baumannii_MDR-TJ 934298 935503 --> 924298 to 945503 CP001937.2_Acinetobacter_baumannii_MDR-ZJ06 306267 305062 --> 295062 to 316267 CP002522.2_Acinetobacter_baumannii_TCDC-AB0715 3194910 3193705 --> 3183705 to 3204910 CP003856.1_Acinetobacter_baumannii_TYTH-1 3248375 3247170 --> 3237170 to 3258375 CP006768.1_Acinetobacter_baumannii_ZW85-1 921662 922867 --> 911662 to 932867 CP000521.1_Acinetobacter_baumannii_ATCC17978 2990721 2989667 and 2944541 2944384 --> 2934384 3000721 -
After click the button "Generate blastn Files", manully perform the command "blastn"
makeblastdb -in 2.easyfig.fa -dbtype nucl blastn -db 2.easyfig.fa -query 1.easyfig.fa -num_threads 15 -outfmt 6 -strand both -evalue 0.1 -max_target_seqs 1 > 12.easyfig.out makeblastdb -in 3.easyfig.fa -dbtype nucl blastn -db 3.easyfig.fa -query 2.easyfig.fa -num_threads 15 -outfmt 6 -strand both -evalue 0.1 -max_target_seqs 1 > 23.easyfig.out makeblastdb -in 4.easyfig.fa -dbtype nucl blastn -db 4.easyfig.fa -query 3.easyfig.fa -num_threads 15 -outfmt 6 -strand both -evalue 0.1 -max_target_seqs 1 > 34.easyfig.out makeblastdb -in 5.easyfig.fa -dbtype nucl blastn -db 5.easyfig.fa -query 4.easyfig.fa -num_threads 15 -outfmt 6 -strand both -evalue 0.1 -max_target_seqs 1 > 45.easyfig.out ... -
Generate as an svg file, add the name of genome manually. Note that the following colors were used.
#https://github.com/mjsull/Easyfig/wiki/Example-2.-whole-genome-comparison normal-minimum: aqua normal-maximum: blue inverted-minimum: orange inverted-maximum: red -
Attachment example3_settings.easycfg
02. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/A.baumannii_AYE.gbk 03. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/A.baumannii_SDF.gbk 04. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/A.baumannii_ACICU.gbk 05. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/Acinetobacter_baumannii_1656-2.gbk 06. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/Acinetobacter_baumannii_AB030.gbk 07. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/Acinetobacter_baumannii_AB031.gbk 08. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/Acinetobacter_baumannii_AB0057.gbk 09. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/Acinetobacter_baumannii_AB307-0294.gbk 10. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/Acinetobacter_baumannii_AbH12O-A2.gbk 11. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/Acinetobacter_baumannii_AC29.gbk 12. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/Acinetobacter_baumannii_BJAB0715.gbk 13. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/Acinetobacter_baumannii_BJAB0868.gbk 14. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/Acinetobacter_baumannii_BJAB07104.gbk 15. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/Acinetobacter_baumannii_D1279779.gbk 16. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/Acinetobacter_baumannii_LAC-4.gbk 17. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/Acinetobacter_baumannii_MDR-TJ.gbk 18. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/Acinetobacter_baumannii_MDR-ZJ06.gbk 19. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/Acinetobacter_baumannii_TCDC-AB0715.gbk 20. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/Acinetobacter_baumannii_TYTH-1.gbk 21. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/Acinetobacter_baumannii_ZW85-1.gbk 01. /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/A.baumannii_ATCC17978.gbk 02 964325 985530 False 03 844002 865207 False 04 2969470 2990675 False 05 3007031 3028236 False 06 1780095 1801300 False 07 3291965 3313170 False 08 3076114 3097319 False 09 964367 985572 False 10 2879346 2900551 False 11 1273600 1294805 False 12 3049750 3070955 False 13 2939770 2960975 False 14 3032398 3053603 False 15 2743929 2765134 False 16 966323 987528 False 17 924298 945503 False 18 295062 316267 False 19 3183705 3204910 False 20 3237170 3258375 False 21 911662 932867 False 01 2934384 3000721 False /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/12.easyfig.out /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/23.easyfig.out /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/34.easyfig.out /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/45.easyfig.out /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/56.easyfig.out /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/67.easyfig.out /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/78.easyfig.out /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/89.easyfig.out /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/910.easyfig.out /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/1011.easyfig.out /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/1112.easyfig.out /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/1213.easyfig.out /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/1314.easyfig.out /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/1415.easyfig.out /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/1516.easyfig.out /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/1617.easyfig.out /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/1718.easyfig.out /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/1819.easyfig.out /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/1920.easyfig.out /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/2021.easyfig.out /mnt/md1/Data_Tam_ABAYE_RS05070_on_A_calcoaceticus_baumannii_complex/comparative_genome_plots/Easyfig_files3/zzzz.bmp Bitmap (bmp) 5000 150 500 centre 1 1 5000 0 0.001 0 0 255 255 #00ffff 255 165 0 #ffa500 0 0 255 #0000ff 255 0 0 #ff0000 1 Top & Bottom None locus_tag 20 2 1 1 64 224 208 #40e0d0 arrow 0 255 140 0 #ff8c00 arrow 0 165 42 42 #a52a2a rect 0 0 191 255 #00bfff rect 0 72 61 139 #483d8b arrow None 0 1000 1000 200 Auto 0 Histogram 1 255 0 0 #FF0000 0 0 255 #0000FF 10 -
identificatio of IS (Insertion Sequence) elements
#extracted sequence segments from the two isolates, specifically: # ATCC19606: 930469 to 951674 — segment1 # ATCC17978: 2,934,384 to 3,000,721 — segment2 #Then, I compared the two segments and found that positions 1-11055 of segment1 mapped to 66338-55284 of segment2, and positions 11049-21206 of segment1 mapped to 10158-23 of segment2. This means the sequence from 10159-55283 of segment2 (about 45 kb nt) is not mapped. I then extracted the 45 kb sequence (see the attached fasta file). I attempted to detect IS elements using the tool ISEScan (https://academic.oup.com/bioinformatics/article/33/21/3340/3930124). Four ISs were detected (see 45kb.fasta.xlsx; for more detailed results, see 45kb.fasta.zip). samtools faidx Acinetobacter_baumannii_ATCC19606.gbk_converted.fna CP059040.1:930469-951674 > ../ATCC19606_segment.fasta samtools faidx A.baumannii_ATCC17978.gbk_converted.fna CP000521.1:2934384-3000721 > ../ATCC17978_segment.fasta makeblastdb -in ATCC17978_segment.fasta -dbtype nucl blastn -db ATCC17978_segment.fasta -query ATCC19606_segment.fasta -num_threads 15 -outfmt 6 -strand both -evalue 0.1 > ATCC19606_segment_on_ATCC17978_segment.blastn samtools faidx ATCC17978_segment.fasta CP000521.1_2934384_3000721:10159-55283 > 45kb.fasta-
ISEScan: Description: Although not a database, ISEScan is a software tool used to identify IS elements in bacterial genome sequences. It can be helpful for researchers looking to analyze newly sequenced genomes for the presence of IS elements. Website: Available on platforms like GitHub for download and integration into bioinformatics workflows.
-
TnCentral including ISFinder: Description: TnCentral is a more comprehensive resource that includes information about transposons, which are larger and more complex than simple IS elements but often contain IS sequences as part of their structure. This database provides detailed information about transposon structures, including associated genes and regulatory features.
-
ISsaga: ISsaga is a web-based tool for the identification and annotation of insertion sequences in prokaryotic genomes. It provides various features for IS element analysis, including detection, classification, and visualization. You can access ISsaga here: ISsaga (http://issaga.biotoul.fr/ISsaga/issaga_index.php)
-
ISFinder: ISFinder is a curated database and analysis platform for insertion sequences in prokaryotic genomes. It provides a comprehensive collection of IS sequences and tools for sequence analysis, classification, and annotation. You can access ISFinder here: ISFinder For ISfinder please cite: Siguier P. et al. (2006) ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34: D32-D36 (link pubmed) and the database URL (http://www-is.biotoul.fr).
-
For ISbrowser please cite: Kichenaradja P. et al. (2010) ISbrowser: an extension of ISfinder for visualizing insertion sequences in prokaryotic genomes. Nucleic Acids Res. 38: D62-D68 (link pubmed). For ISsaga please cite: Varani A. et al. (2011) ISsaga: an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes, Genome Biology 2011, 12:R30 (link pubmed).
-
ISMapper: ISMapper is a tool for mapping insertion sequences in bacterial genomes. It uses paired-end sequence data to identify IS element insertion sites and provides information about their genomic context. You can access ISMapper here: ISMapper ISMapper: identifying transposase insertion sites in bacterial genomes from short read sequence data https://pubmed.ncbi.nlm.nih.gov/26336060/
-
ISseeker: ISseeker is a software package for the identification and annotation of insertion sequences in bacterial genomes. It provides a user-friendly interface for IS element detection and characterization. You can access ISseeker here: ISseeker
-
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Kraken2 Installation and Usage Guide
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Should the inputs for GSVA be normalized or raw?
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- Risks of Rebooting into Rescue Mode
- 足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python