How does the adapter in Illumina sequencing work?
gene_x 0 like s 1871 view s
Tags: technique, sequencing
Why are adapter sequences trimmed from only the 3' ends of reads? https://support.illumina.com.cn/bulletins/2016/04/adapter-trimming-why-are-adapter-sequences-trimmed-from-only-the--ends-of-reads.html
Expands one nucleic base at a time https://www.researchgate.net/figure/Illumina-sequencing-process-A-DNA-library-Breaks-the-genome-DNA-to-form-DNA_fig3_357155980
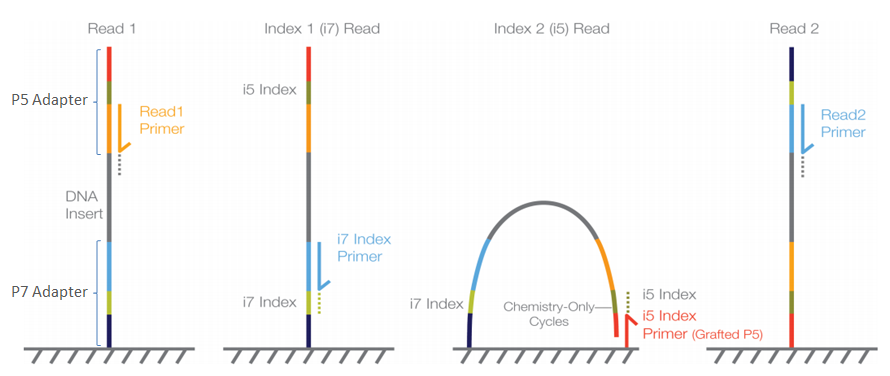
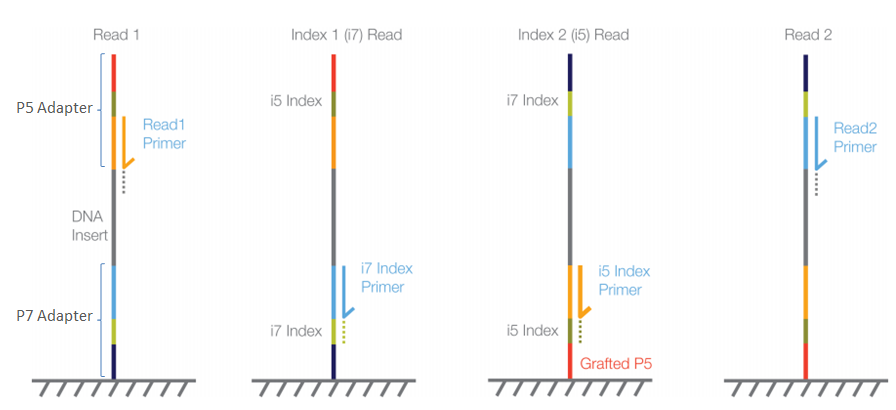
In Illumina sequencing, the barcode (also known as the index) is indeed a critical part of the sequencing process because it allows for the identification and demultiplexing of multiple samples that are sequenced together in the same run.
Here’s how it works:
-
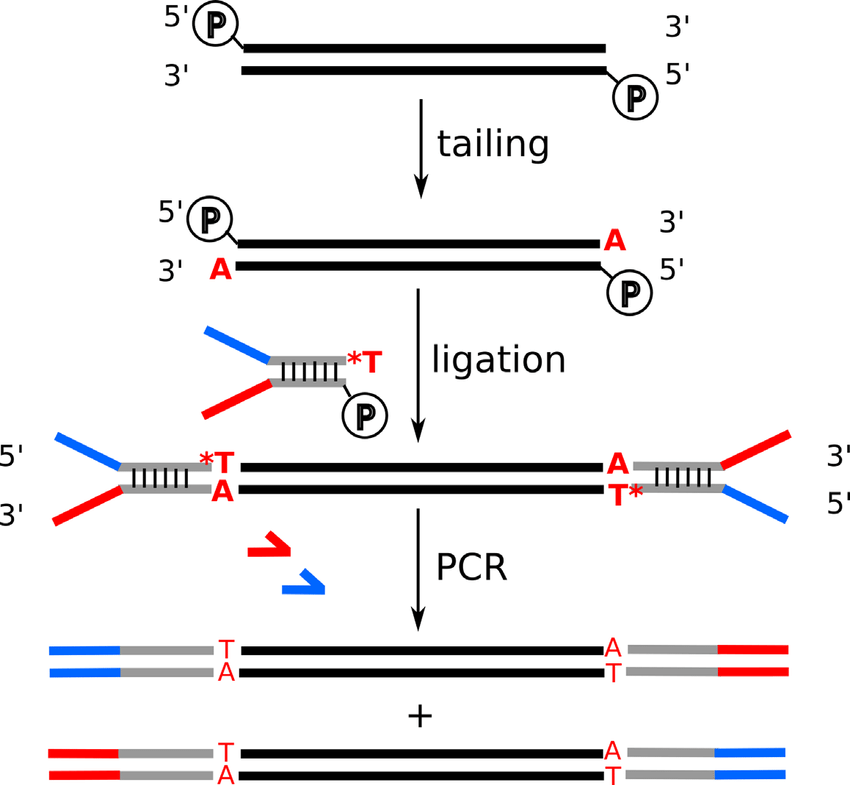
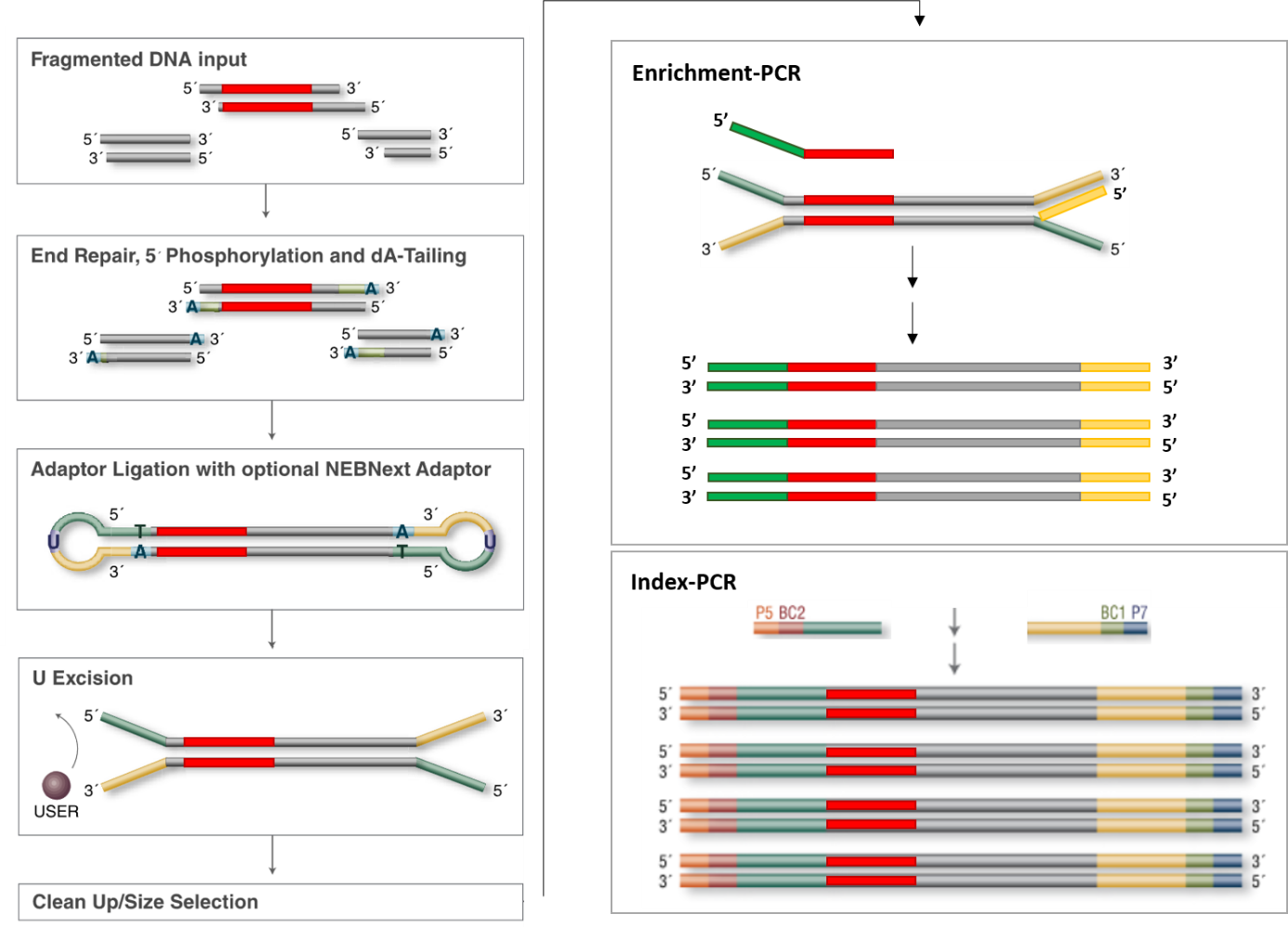
Adapter Ligation: First, adapters are ligated to the fragmented DNA. These adapters contain the sequences for P5 and P7 priming sites, necessary for flow cell attachment and the initiation of the sequencing reaction.
-
Index Sequences: The adapters also include index sequences (barcodes). In the case of dual-indexing, one index (Index 1) is on the adapter ligated at the P7 end, and another index (Index 2) is on the adapter ligated at the P5 end. These indexes are unique to each sample.
-
Sequencing Initiation: Sequencing begins with the binding of sequencing primers to their complementary sites on the adapters—not directly from the index sequences. However, the index sequences are read during specific additional sequencing reactions:
* For Read 1, sequencing starts from the P5 end. * If performing paired-end sequencing, after Read 1 is complete, the machine performs a read of the Index 1 sequence. * Then, the flow cell is reconfigured to sequence Read 2 from the P7 end. * Finally, if dual-indexing, the Index 2 sequence is read. -
Index Reading (Read1 Primer and i7 Index Primer): The indexes are not part of the main sequence reads (Read 1 or Read 2) but are read in separate, dedicated sequencing reactions using specific index primers after the completion of the standard sequencing cycles.
The crucial point is that the sequencing of the index sequences happens after the main DNA fragment has been sequenced, during dedicated index read cycles. The readout of the indexes is integral to the sequencing run and allows the software to assign each sequence to the correct sample in the analysis phase, enabling the pooling of multiple samples in a single sequencing run. This process is called demultiplexing.
Tn5 adapter https://teichlab.github.io/scg_lib_structs/methods_html/plate_and_piATAC-seq.html
Y-shaped-adaptors https://www.researchgate.net/figure/DNA-template-ligation-with-Y-shaped-adaptors-Blunt-ended-ds-DNA-templates-5_fig2_323640739
(0) Final library structure:
5'- AATGATACGGCGACCACCGAGATCTACACNNNNNNNNTCGTCGGCAGCGTCAGATGTGTATAAGAGACAGXXXXXXXX...XXXXXXXXCTGTCTCTTATACACATCTCCGAGCCCACGAGACNNNNNNNNATCTCGTATGCCGTCTTCTGCTTG
TTACTATGCCGCTGGTGGCTCTAGATGTGNNNNNNNNAGCAGCCGTCGCAGTCTACACATATTCTCTGTCXXXXXXXX...XXXXXXXXGACAGAGAATATGTGTAGAGGCTCGGGTGCTCTGNNNNNNNNTAGAGCATACGGCAGAAGACGAAC -5'
Illumina P5 i5 s5 ME cDNA ME s7 i7 Illumina P7
Library sequencing:
(1) Add read 1 sequencing primer to sequence the first read (bottom strand as template):
Primer1
5'- TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG|--READ1---->
3'- TTACTATGCCGCTGGTGGCTCTAGATGTGNNNNNNNNAGCAGCCGTCGCAGTCTACACATATTCTCTGTCXXXXXXXX...XXXXXXXXGACAGAGAATATGTGTAGAGGCTCGGGTGCTCTGNNNNNNNNTAGAGCATACGGCAGAAGACGAAC -5'
(2) Add index 1 sequencing primer to sequence the first index (i7) (bottom strand as template, 8 cycles):
5'- CTGTCTCTTATACACATCTCCGAGCCCACGAGAC------>
3'- TTACTATGCCGCTGGTGGCTCTAGATGTGNNNNNNNNAGCAGCCGTCGCAGTCTACACATATTCTCTGTCXXXXXXXX...XXXXXXXXGACAGAGAATATGTGTAGAGGCTCGGGTGCTCTGNNNNNNNNTAGAGCATACGGCAGAAGACGAAC -5'
(3) Cluster regeneration, add Index 2 sequencing primer to sequence the second index (i5) (top strand as template, 8 cycles. Single cells can be identified as the combination of i5 and i7):
5'- AATGATACGGCGACCACCGAGATCTACACNNNNNNNNTCGTCGGCAGCGTCAGATGTGTATAAGAGACAGXXXXXXXX...XXXXXXXXCTGTCTCTTATACACATCTCCGAGCCCACGAGACNNNNNNNNATCTCGTATGCCGTCTTCTGCTTG
<-------AGCAGCCGTCGCAGTCTACACATATTCTCTGTC -5'
(4) Add read 2 sequencing primer to sequence the second read (top strand as template):
5'- AATGATACGGCGACCACCGAGATCTACACNNNNNNNNTCGTCGGCAGCGTCAGATGTGTATAAGAGACAGXXXXXXXX...XXXXXXXXCTGTCTCTTATACACATCTCCGAGCCCACGAGACNNNNNNNNATCTCGTATGCCGTCTTCTGCTTG
<----READ2--|GACAGAGAATATGTGTAGAGGCTCGGGTGCTCTG -5'
Primer2
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Kraken2 Installation and Usage Guide
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Should the inputs for GSVA be normalized or raw?
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- Risks of Rebooting into Rescue Mode
- 足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
重新审视诊断:微生物细胞游离DNA测序:解决与植入物相关的心血管感染中的未解决挑战
Native elongating transcript sequencing (NET-seq)