Step-by-Step Analysis of Transposon Insertion Sites
gene_x 0 like s 1196 view s
Tags: sequencing, pipeline
-
The script processes paired-end sequencing data from a SAM file to identify reads mapping to a specific reference and position, outputting the results in FASTQ format. It imports necessary Biopython libraries, defines a function to parse the CIGAR string, and includes a function to save sequences as FASTQ files. The main function, extract_reads, extracts read pairs from the SAM file that map to a given reference and position, checking for soft-clipped sequences indicative of transposon insertions. It categorizes the reads into original, bacterial, and transposon sequences, and saves them in separate FASTQ files. The script concludes by printing the counts of each type of read pair saved. (Need to be debugged!)
python3 check_insertion_site_with_transposon16.py import re from Bio import SeqIO from Bio.Seq import Seq from Bio.SeqRecord import SeqRecord def parse_cigar(cigar): """ Parse the CIGAR string and return a list of tuples (operation, length). """ return [(int(length), op) for length, op in re.findall(r'(\d+)([MIDNSHP=X])', cigar)] def save_fastq(sequences, file_name, read_ids): """ Save sequences to a FASTQ file. """ records = [SeqRecord(Seq(seq), id=read_id, description="", letter_annotations={"phred_quality": [40]*len(seq)}) for seq, read_id in zip(sequences, read_ids)] SeqIO.write(records, file_name, "fastq") def extract_reads(sam_file, reference, position): """ Extract paired reads mapping to a specific position and categorize them. """ original_reads_R1 = [] original_reads_R2 = [] bacterial_reads_R1 = [] bacterial_reads_R2 = [] transposon_reads_R1 = [] transposon_reads_R2 = [] read_ids_R1 = [] read_ids_R2 = [] read_pairs = {} # To store read pairs with open(sam_file, 'r') as file: for line in file: if line.startswith('@'): continue parts = line.split('\t') flag = int(parts[1]) ref_name = parts[2] start_pos = int(parts[3]) cigar = parts[5] seq = parts[9] read_name = parts[0] # Skip unmapped reads if ref_name == '*' or cigar == '*': continue # Skip reads not mapped to the specified reference if ref_name != reference: continue # Determine if the read is R1 or R2 based on the flag is_read1 = flag & 64 != 0 is_read2 = flag & 128 != 0 # Parse the CIGAR string parsed_cigar = parse_cigar(cigar) # Calculate the end position of the aligned part of the read aligned_length = sum(length for length, op in parsed_cigar if op in "M=X") end_pos = start_pos + aligned_length - 1 # Store the read and its pair if read_name not in read_pairs: read_pairs[read_name] = [None, None] # [R1, R2] if is_read1: read_pairs[read_name][0] = (seq, start_pos, end_pos, parsed_cigar, read_name) if is_read2: read_pairs[read_name][1] = (seq, start_pos, end_pos, parsed_cigar, read_name) for read_name, pair in read_pairs.items(): if pair[0] and pair[1]: read1, read2 = pair # Check if one of the reads contains a soft-clip has_soft_clip1 = any(op == 'S' for length, op in read1[3]) has_soft_clip2 = any(op == 'S' for length, op in read2[3]) if has_soft_clip1 or has_soft_clip2: original_reads_R1.append(read1[0]) original_reads_R2.append(read2[0]) read_ids_R1.append(read1[4] + "/1") read_ids_R2.append(read2[4] + "/2") # Extract bacterial parts only bacterial_seq1 = ''.join([read1[0][i:i+length] for i, (length, op) in enumerate(read1[3]) if op in "M=X"]) bacterial_seq2 = ''.join([read2[0][i:i+length] for i, (length, op) in enumerate(read2[3]) if op in "M=X"]) bacterial_reads_R1.append(bacterial_seq1) bacterial_reads_R2.append(bacterial_seq2) # Extract transposon parts only (soft-clipped parts) transposon_seq1 = ''.join([read1[0][i:i+length] for i, (length, op) in enumerate(read1[3]) if op == 'S']) transposon_seq2 = ''.join([read2[0][i:i+length] for i, (length, op) in enumerate(read2[3]) if op == 'S']) transposon_reads_R1.append(transposon_seq1) transposon_reads_R2.append(transposon_seq2) save_fastq(original_reads_R1, 'original_reads_R1.fastq', read_ids_R1) save_fastq(original_reads_R2, 'original_reads_R2.fastq', read_ids_R2) save_fastq(bacterial_reads_R1, 'bacterial_reads_R1.fastq', read_ids_R1) save_fastq(bacterial_reads_R2, 'bacterial_reads_R2.fastq', read_ids_R2) save_fastq(transposon_reads_R1, 'transposon_reads_R1.fastq', read_ids_R1) save_fastq(transposon_reads_R2, 'transposon_reads_R2.fastq', read_ids_R2) print(f"Total original read pairs saved: {len(original_reads_R1)}") print(f"Total bacterial read pairs saved: {len(bacterial_reads_R1)}") print(f"Total transposon read pairs saved: {len(transposon_reads_R1)}") return len(original_reads_R1), len(bacterial_reads_R1), len(transposon_reads_R1) # Example usage sam_file = 'initial_mutants_rep1_no_insertion_sites.sam' reference = 'NZ_AKKR01000009' position = 31537 original_pairs_count, bacterial_pairs_count, transposon_pairs_count = extract_reads(sam_file, reference, position) print(f"Number of original read pairs: {original_pairs_count}") print(f"Number of bacterial read pairs: {bacterial_pairs_count}") print(f"Number of transposon read pairs: {transposon_pairs_count}") -
Reversing Complementing FASTQ Reads. This effectively swaps and reverse complements the read pairs.
seqtk seq -r original_reads_R1.fastq > original_reads_rc_R2.fastq seqtk seq -r original_reads_R2.fastq > original_reads_rc_R1.fastq -
Counting Specific Sequences in FASTQ Files
grep "CCTACAACAAAGCTCTCATCAAC" original_reads_rc_R1.fastq | wc -l #19186 grep "CCTACAACAAAGCTCTCATCAAC" original_reads_rc_R2.fastq | wc -l #42069 -
Running Transposon Position Profiling (TPP)
#This command runs the tpp (Transposon Position Profiling) tool, which is used for mapping transposon insertions. # -bwa /usr/bin/bwa: Specifies the path to the BWA alignment tool. # -protocol Tn5: Specifies the protocol used, in this case, Tn5 transposon. # -ref contig_1_1.fna: Specifies the reference genome file. # -reads1 original_reads_rc_R1.fastq -reads2 original_reads_rc_R2.fastq: Specifies the input FASTQ files. # -output contig_1_1_rc: Specifies the output prefix. # -primer CCTACAACAAAGCTCTCATCAAC: Specifies the primer sequence used for mapping. # -mismatches 1: Allows up to 1 mismatch in the alignment. # -bwa-alg mem: Uses the mem algorithm in BWA. # -replicon-ids contig_1_1: Specifies the replicon IDs. python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref contig_1_1.fna -reads1 original_reads_rc_R1.fastq -reads2 original_reads_rc_R2.fastq -output contig_1_1_rc -primer CCTACAACAAAGCTCTCATCAAC -mismatches 1 -bwa-alg mem -replicon-ids contig_1_1 mv tpp.cfg contig_1_1_rc.tpp.cfg # TA_sites: 84291 # TAs_hit: 54 # density: 0.001 # max_count (among templates): 1 # max_site (coordinate): 84038 #This command runs tpp again but only uses original_reads_rc_R1.fastq. #Analysis: Indicates an issue because BWA alignments expected read2 to be from the bacterial genome, which #was not the case. python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref contig_1_1.fna -reads1 original_reads_rc_R1.fastq -output contig_1_1_rc_only_reads1 -primer CCTACAACAAAGCTCTCATCAAC -mismatches 1 -bwa-alg mem -replicon-ids contig_1_1 mv tpp.cfg contig_1_1_rc_only_reads1.tpp.cfg #Max_count (among templates): 2013 #Max_site (coordinate): 31531 #This command runs tpp again but only uses original_reads_rc_R2.fastq. python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref contig_1_1.fna -reads1 original_reads_rc_R2.fastq -output contig_1_1_rc_only_reads2 -primer CCTACAACAAAGCTCTCATCAAC -mismatches 1 -bwa-alg mem -replicon-ids contig_1_1 mv tpp.cfg contig_1_1_rc_only_reads2.tpp.cfg #Max_count (among templates): 7210 #Max_site (coordinate): 31531 -
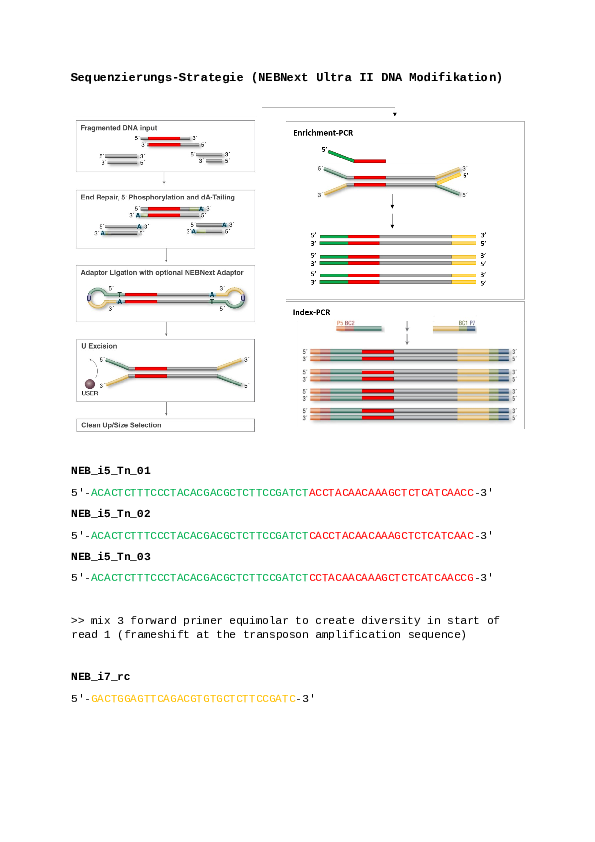
Conclusion: With the pipeline described above, we aim to identify the read coverage at the first junction site between the bacterial genome and the transposon (see Figure_1.png). By default, we focus only on the second junction site between the transposon and the bacterial genome. For an example of how to identify these junction sites, see the following command.
#-primer AGATGTGTATAAGAGACAG -mismatches 1
#-primer TAAGAGACAG -mismatches 1
#mismatches1 1
#primer_start_window 0,159
#window_size -1
#transposon Tn5
#protocol Tn5
python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m.fna -reads1 ./240405_VH00358_89_AAFC5MTM5/kr1/initial_mutants_rep1_S25_R1_001.fastq -reads2 ./240405_VH00358_89_AAFC5MTM5/kr1/initial_mutants_rep1_S25_R2_001.fastq -output initial_mutants_rep1 -primer TAAGAGACAG -mismatches 1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689
mv tpp.cfg initial_mutants_rep1.tpp.cfg
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Kraken2 Installation and Usage Guide
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Should the inputs for GSVA be normalized or raw?
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- Risks of Rebooting into Rescue Mode
- 足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
piRNA 2024 Ute from raw counts
Tn-seq analysis pipeline (improved)
GSVA calculation for NanoString data
How to use H3K27ac, H3K4me1, and RNA-seq to identify enhancers and their target genes?