Tn-seq analysis pipeline (improved3)
gene_x 0 like s 877 view s
Tags: pipeline
only one mutant, PacBio long-sequencing find the positions of transposons
Häufigkeit in mixed mutants using short-sequencing
DEGs regarding Häufigkeit using short-sequencing
-
Overview of Data Processing Procedure
1. Convert .fastq files to .fasta format (.reads). "AGCTTCAGGGTTGAGATGTGTATAAGAGACAG", allowed a mismatch of 1 nt 2. Identify reads with the transposon prefix in R1. The sequence searched for is "AGCTTCAGGGTTGAGATGTGTATAAGAGACAG", allowed a mismatch of 1 nt, which must start between cycles 5 and 10 (inclusive). (Note that this ends in the canonical terminus of the Himar1 transposon, TGTTA.) The “staggered” position of this sequence is due to insertion a few nucleotides of variable length in the primers used in the Tn-Seq sample prep protocol (e.g. 4 variants of Sol_AP1_57, etc.). The number of mimatches allowed in searching reads for the transposon sequence pattern can be adjusted as an option in the interface; the default is 1. #What are TACCACGACCA? 3. Extract genomic part of read 1. This is the suffix following the transposon sequence pattern above. However, for reads coming from fragments shorter than the read length, the adapter might appear at the other end of R1, TACCACGACCA. If so, the adapter suffix is stripped off. (These are referred to as “truncated” reads, but they can still be mapped into the genome just fine by BWA.) The length of the genomic part must be at least 20 bp. 3. Extract barcodes from read 2. Read 2 is searched for GATGGCCGGTGGATTTGTGnnnnnnnnnnTGGTCGTGGTAT”. The length of the barcode is typically 10 bp, but can be varaible, and must be between 5-15 bp. 4. Extract genomic portions of read 2. This is the part following TGGTCGTGGTAT…. It is often the whole suffix of the read. However, if the read comes from a short DNA fragment that is shorter than the read length, the adapter on the other end might appear, in which case it is stripped off and the nucleotides in the middle representing the genomic insert, TGGTCGTGGTATxxxxxxxTAACAGGTTGGCTGATAAG. The insert must be at least 20 bp long (inserts shorter than this are discarded, as they might map to spurious locations in the genome). 5. Map genomic parts of R1 and R2 into the genome using BWA. Mismatches are allowed, but indels are ignored. No trimming is performed. BWA is run in ‘sampe’ mode (treating reads as pairs). Both reads of a pair must map (on opposite strands) to be counted. 6. Count the reads mapping to each TA site in the reference genome (or all sites for Tn5). 7. Reduce raw read counts to unique template counts. Group reads by barcode AND mapping location of read 2 (aka fragment “endpoints”). 8. Output template counts at each TA site in a .wig file. 9. Calculate statistics like insertion_density and NZ_mean. Look for the site with the max template count. Look for reads matching the primer or vector sequences. -
quality control
./240606_VH00358_96_AAFCFJGM5/kr11/initial_mutants_a_2_S6_R1_001.fastq.gz ./240606_VH00358_96_AAFCFJGM5/kr11/initial_mutants_a_2_S6_R2_001.fastq.gz ./240606_VH00358_96_AAFCFJGM5/kr13/LB_culture_a_2_S7_R1_001.fastq.gz ./240606_VH00358_96_AAFCFJGM5/kr13/LB_culture_a_2_S7_R2_001.fastq.gz ./240606_VH00358_96_AAFCFJGM5/kr15/growthout_control_24h_a_2_S8_R1_001.fastq.gz ./240606_VH00358_96_AAFCFJGM5/kr15/growthout_control_24h_a_2_S8_R2_001.fastq.gz ./240606_VH00358_96_AAFCFJGM5/kr17/extracellular_mutants_24h_a_2_S9_R1_001.fastq.gz ./240606_VH00358_96_AAFCFJGM5/kr17/extracellular_mutants_24h_a_2_S9_R2_001.fastq.gz ./240606_VH00358_96_AAFCFJGM5/kr19/intracellular_mutants_24h_a_2_S10_R1_001.fastq.gz ./240606_VH00358_96_AAFCFJGM5/kr19/intracellular_mutants_24h_a_2_S10_R2_001.fastq.gz #from fastqc results of initial_mutants 49821406 35-161 49821406 35-161 https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=1194086 https://www.ncbi.nlm.nih.gov/Traces/wgs/AKKR01? https://www.ncbi.nlm.nih.gov/Traces/wgs/AKKR01?display=download #Found 4,131 proteins -
modify the tpp scripts
vim ~/.local/lib/python3.10/site-packages/pytpp/tpp_tools.py #search for "DEBUG" #-maxreads 10000 or not_given for take all! #-primer AGATGTGTATAAGAGACAG the default primer of Tn5 is TAAGAGACAG! #-primer-start-window 0,159 set 0,159 as default! #delete to import barcode-file, since we already demultipled the file! # pattern for read 2... # TAGTGGATGATGGCCGGTGGATTTGTG GTAATTACCA TGGTCGTGGTAT CCCAGCGCGACTTCTTCGGCGCACACACC TAACAGGTTGGCTGATAAGTCCCCG?AGAT AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGT # -----const1---------------- --barcode- ---const2--- ------genomic---------------- ------const3-------------------------------------------------------------- -
run Transposon Position Profiling (TPP) on multiple contigs
#https://transit.readthedocs.io/en/latest/transit_running.html; #https://orca2.tamu.edu/tom/iLab.html # Break-down of total reads (49821406): # 29481783 reads (59.2%) lack the expected Tn prefix # Break-down of trimmed reads with valid Tn prefix (20339623): #primer AGCTTCAGGGTTGAGATGTGTATAAGAGACAG # primer_matches: 0 reads (0.0%) contain CTAGAGGGCCCAATTCGCCCTATAGTGAGT (Himar1) # vector_matches: 0 reads (0.0%) contain CTAGACCGTCCAGTCTGGCAGGCCGGAAAC (phiMycoMarT7) # adapter_matches: 0 reads (0.0%) contain GATCGGAAGAGCACACGTCTGAACTCCAGTCAC (Illumina/TruSeq index) # misprimed_reads: 0 reads (0.0%) contain Himar1 prefix but don't end in TGTTA #kr11.trimmed1_failed_trim 22072406 #-rw-rw-r-- 1 jhuang jhuang 2,7G Jun 11 15:44 kr11.trimmed1 20339623 #-rw-rw-r-- 1 jhuang jhuang 2,9G Jun 11 15:46 kr11.trimmed2 20339623 #cat initial_mutants_a_2_S6_R1_001.fastq | echo $((`wc -l` / 4)) 49821406=29481783 reads (59.2%) + 20339623 = 49821406 #vs. # Break-down of total reads (49821406): # 29356859 reads (58.9%) lack the expected Tn prefix # Break-down of trimmed reads with valid Tn prefix (20464547): # primer_matches: 0 reads (0.0%) contain CTAGAGGGCCCAATTCGCCCTATAGTGAGT (Himar1) # vector_matches: 0 reads (0.0%) contain CTAGACCGTCCAGTCTGGCAGGCCGGAAAC (phiMycoMarT7) # adapter_matches: 0 reads (0.0%) contain GATCGGAAGAGCACACGTCTGAACTCCAGTCAC (Illumina/TruSeq index) # misprimed_reads: 0 reads (0.0%) contain Himar1 prefix but don't end in TGTTA # read_length: 100 bp # mean_R1_genomic_length: 73.4 bp # mean_R2_genomic_length: 86.4 bp #conda deactivate ##Test AGCTTCAGGGTTGAGATGTGTATAAGAGACAG --> TAAGAGACAG, the results are similar! # Note that "-primer-start-window 0,161" is invalid and cannot replace the default value! #python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m_.fasta -reads1 240606_VH00358_96_AAFCFJGM5/kr11/initial_mutants_a_2_S6_R1_001.fastq.gz -reads2 240606_VH00358_96_AAFCFJGM5/kr11/initial_mutants_a_2_S6_R2_001.fastq.gz -output kr11_10nt_primer -primer TAAGAGACAG -mismatches 1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689 #mv tpp.cfg kr11_10nt_primer_tpp.cfg #for initial_mutants python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m_.fasta -reads1 240606_VH00358_96_AAFCFJGM5/kr11/initial_mutants_a_2_S6_R1_001.fastq.gz -reads2 240606_VH00358_96_AAFCFJGM5/kr11/initial_mutants_a_2_S6_R2_001.fastq.gz -output initial_mutants -primer AGCTTCAGGGTTGAGATGTGTATAAGAGACAG -mismatches 1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689 mv tpp.cfg initial_mutants_tpp.cfg #primer_start_window 0,159 # Break-down of total reads (49821406): # 29481783 reads (59.2%) lack the expected Tn prefix # Break-down of trimmed reads with valid Tn prefix (20339623): # primer_matches: 0 reads (0.0%) contain CTAGAGGGCCCAATTCGCCCTATAGTGAGT (Himar1) # vector_matches: 0 reads (0.0%) contain CTAGACCGTCCAGTCTGGCAGGCCGGAAAC (phiMycoMarT7) # adapter_matches: 0 reads (0.0%) contain GATCGGAAGAGCACACGTCTGAACTCCAGTCAC (Illumina/TruSeq index) # misprimed_reads: 0 reads (0.0%) contain Himar1 prefix but don't end in TGTTA #for LB_culture python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m_.fasta -reads1 ./240606_VH00358_96_AAFCFJGM5/kr13/LB_culture_a_2_S7_R1_001.fastq.gz -reads2 ./240606_VH00358_96_AAFCFJGM5/kr13/LB_culture_a_2_S7_R2_001.fastq.gz -output LB_culture -primer AGCTTCAGGGTTGAGATGTGTATAAGAGACAG -mismatches 1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689 mv tpp.cfg LB_culture_tpp.cfg #for growthout_control_24h python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m_.fasta -reads1 ./240606_VH00358_96_AAFCFJGM5/kr15/growthout_control_24h_a_2_S8_R1_001.fastq.gz -reads2 ./240606_VH00358_96_AAFCFJGM5/kr15/growthout_control_24h_a_2_S8_R2_001.fastq.gz -output growthout_control_24h -primer AGCTTCAGGGTTGAGATGTGTATAAGAGACAG -mismatches 1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689 mv tpp.cfg growthout_control_24h_tpp.cfg #for extracellular_mutants_24h python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m_.fasta -reads1 ./240606_VH00358_96_AAFCFJGM5/kr17/extracellular_mutants_24h_a_2_S9_R1_001.fastq.gz -reads2 ./240606_VH00358_96_AAFCFJGM5/kr17/extracellular_mutants_24h_a_2_S9_R2_001.fastq.gz -output extracellular_mutants_24h -primer AGCTTCAGGGTTGAGATGTGTATAAGAGACAG -mismatches 1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689 mv tpp.cfg extracellular_mutants_24h_tpp.cfg #for intracellular_mutants_24h python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m_.fasta -reads1 ./240606_VH00358_96_AAFCFJGM5/kr19/intracellular_mutants_24h_a_2_S10_R1_001.fastq.gz -reads2 ./240606_VH00358_96_AAFCFJGM5/kr19/intracellular_mutants_24h_a_2_S10_R2_001.fastq.gz -output intracellular_mutants_24h -primer AGCTTCAGGGTTGAGATGTGTATAAGAGACAG -mismatches 1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689 mv tpp.cfg intracellular_mutants_24h_tpp.cfg -
generate statistics tables in Excel-format from the multiple contig running.
for sample in initial_mutants LB_culture growthout_control_24h extracellular_mutants_24h intracellular_mutants_24h; do echo "cd ${sample}" echo "cp ${sample}.tn_stats ${sample}.tn_stats_" echo "#Delete all general statistics before the table data in ${sample}.tn_stats_; delete the content after \"# FR_corr (Fwd templates vs. Rev templates):\"" echo "sed -i 's/read_count (TA sites only, for Himar1)/read_counts/g' *.tn_stats_" echo "sed -i 's/NZ_mean (among templates)/NZ_mean (mean template count over non-zero TA sites)/g' *.tn_stats_" echo "python3 ~/Scripts/parse_tn_stats.py ${sample}.tn_stats_ ${sample}.tn_stats.xlsx" echo "#calculate the sum of the first and second columns by \"=SUM(B2:B130)\" and \"=SUM(C2:C130)\"" echo "mkdir ${sample}_wig" echo "mv *.wig ${sample}_wig/" echo "zip -r ${sample}_wig.zip ${sample}_wig/" done cd initial_mutants cp initial_mutants.tn_stats initial_mutants.tn_stats_ #Delete all general statistics before the table data in initial_mutants.tn_stats_; delete the content after "# FR_corr (Fwd templates vs. Rev templates):" sed -i 's/read_count (TA sites only, for Himar1)/read_counts/g' *.tn_stats_ sed -i 's/NZ_mean (among templates)/NZ_mean (mean template count over non-zero TA sites)/g' *.tn_stats_ python3 ~/Scripts/parse_tn_stats.py initial_mutants.tn_stats_ initial_mutants.tn_stats.xlsx #calculate the sum of the first and second columns by "=SUM(B2:B130)" and "=SUM(C2:C130)" #16,228,513 and 2,454,346 cd LB_culture cp LB_culture.tn_stats LB_culture.tn_stats_ #Delete all general statistics before the table data in LB_culture.tn_stats_; delete the content after "# FR_corr (Fwd templates vs. Rev templates):" sed -i 's/read_count (TA sites only, for Himar1)/read_counts/g' *.tn_stats_ sed -i 's/NZ_mean (among templates)/NZ_mean (mean template count over non-zero TA sites)/g' *.tn_stats_ python3 ~/Scripts/parse_tn_stats.py LB_culture.tn_stats_ LB_culture.tn_stats.xlsx #calculate the sum of the first and second columns by "=SUM(B2:B130)" and "=SUM(C2:C130)" #19735541, 3266320 cd growthout_control_24h cp growthout_control_24h.tn_stats growthout_control_24h.tn_stats_ #Delete all general statistics before the table data in growthout_control_24h.tn_stats_; delete the content after "# FR_corr (Fwd templates vs. Rev templates):" sed -i 's/read_count (TA sites only, for Himar1)/read_counts/g' *.tn_stats_ sed -i 's/NZ_mean (among templates)/NZ_mean (mean template count over non-zero TA sites)/g' *.tn_stats_ python3 ~/Scripts/parse_tn_stats.py growthout_control_24h.tn_stats_ growthout_control_24h.tn_stats.xlsx #calculate the sum of the first and second columns by "=SUM(B2:B130)" and "=SUM(C2:C130)" #23812866, 3487969 cd extracellular_mutants_24h cp extracellular_mutants_24h.tn_stats extracellular_mutants_24h.tn_stats_ #Delete all general statistics before the table data in extracellular_mutants_24h.tn_stats_; delete the content after "# FR_corr (Fwd templates vs. Rev templates):" sed -i 's/read_count (TA sites only, for Himar1)/read_counts/g' *.tn_stats_ sed -i 's/NZ_mean (among templates)/NZ_mean (mean template count over non-zero TA sites)/g' *.tn_stats_ python3 ~/Scripts/parse_tn_stats.py extracellular_mutants_24h.tn_stats_ extracellular_mutants_24h.tn_stats.xlsx #calculate the sum of the first and second columns by "=SUM(B2:B130)" and "=SUM(C2:C130)" #6491071, 236041 cd intracellular_mutants_24h cp intracellular_mutants_24h.tn_stats intracellular_mutants_24h.tn_stats_ #Delete all general statistics before the table data in intracellular_mutants_24h.tn_stats_; delete the content after "# FR_corr (Fwd templates vs. Rev templates):" sed -i 's/read_count (TA sites only, for Himar1)/read_counts/g' *.tn_stats_ sed -i 's/NZ_mean (among templates)/NZ_mean (mean template count over non-zero TA sites)/g' *.tn_stats_ python3 ~/Scripts/parse_tn_stats.py intracellular_mutants_24h.tn_stats_ intracellular_mutants_24h.tn_stats.xlsx #calculate the sum of the first and second columns by "=SUM(B2:B130)" and "=SUM(C2:C130)" #20619934, 1402849 mkdir initial_mutants_wig mv *.wig initial_mutants_wig/ cd initial_mutants_wig/ python3 ~/Scripts/update_wig_initial_mutants.py cd .. zip -r initial_mutants_wig.zip initial_mutants_wig/ mkdir LB_culture_wig mv *.wig LB_culture_wig/ cd LB_culture_wig/ python3 ~/Scripts/update_wig_LB_culture.py cd .. zip -r LB_culture_wig.zip LB_culture_wig/ mkdir growthout_control_24h_wig mv *.wig growthout_control_24h_wig/ cd growthout_control_24h_wig/ python3 ~/Scripts/update_wig_growthout_control_24h.py cd .. zip -r growthout_control_24h_wig.zip growthout_control_24h_wig/ mkdir extracellular_mutants_24h_wig mv *.wig extracellular_mutants_24h_wig/ cd extracellular_mutants_24h_wig/ python3 ~/Scripts/update_wig_extracellular_mutants_24h.py cd .. zip -r extracellular_mutants_24h_wig.zip extracellular_mutants_24h_wig/ mkdir intracellular_mutants_24h_wig mv *.wig intracellular_mutants_24h_wig/ cd intracellular_mutants_24h_wig/ python3 ~/Scripts/update_wig_intracellular_mutants_24h.py cd .. zip -r intracellular_mutants_24h_wig.zip intracellular_mutants_24h_wig/ zip -r genbank_files.zip genbank_files -
run Transposon Position Profiling (TPP) on merged_genome
#for initial_mutants python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref merged_genome.fasta -reads1 240606_VH00358_96_AAFCFJGM5/kr11/initial_mutants_a_2_S6_R1_001.fastq.gz -reads2 240606_VH00358_96_AAFCFJGM5/kr11/initial_mutants_a_2_S6_R2_001.fastq.gz -output initial_mutants_run2 -primer AGCTTCAGGGTTGAGATGTGTATAAGAGACAG -mismatches 1 -bwa-alg mem mv tpp.cfg initial_mutants_tpp_run2.cfg #for LB_culture python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref merged_genome.fasta -reads1 ./240606_VH00358_96_AAFCFJGM5/kr13/LB_culture_a_2_S7_R1_001.fastq.gz -reads2 ./240606_VH00358_96_AAFCFJGM5/kr13/LB_culture_a_2_S7_R2_001.fastq.gz -output LB_culture_run2 -primer AGCTTCAGGGTTGAGATGTGTATAAGAGACAG -mismatches 1 -bwa-alg mem mv tpp.cfg LB_culture_tpp_run2.cfg #for growthout_control_24h python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref merged_genome.fasta -reads1 ./240606_VH00358_96_AAFCFJGM5/kr15/growthout_control_24h_a_2_S8_R1_001.fastq.gz -reads2 ./240606_VH00358_96_AAFCFJGM5/kr15/growthout_control_24h_a_2_S8_R2_001.fastq.gz -output growthout_control_24h_run2 -primer AGCTTCAGGGTTGAGATGTGTATAAGAGACAG -mismatches 1 -bwa-alg mem mv tpp.cfg growthout_control_24h_tpp_run2.cfg #for extracellular_mutants_24h python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref merged_genome.fasta -reads1 ./240606_VH00358_96_AAFCFJGM5/kr17/extracellular_mutants_24h_a_2_S9_R1_001.fastq.gz -reads2 ./240606_VH00358_96_AAFCFJGM5/kr17/extracellular_mutants_24h_a_2_S9_R2_001.fastq.gz -output extracellular_mutants_24h_run2 -primer AGCTTCAGGGTTGAGATGTGTATAAGAGACAG -mismatches 1 -bwa-alg mem mv tpp.cfg extracellular_mutants_24h_tpp_run2.cfg #for intracellular_mutants_24h python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref merged_genome.fasta -reads1 ./240606_VH00358_96_AAFCFJGM5/kr19/intracellular_mutants_24h_a_2_S10_R1_001.fastq.gz -reads2 ./240606_VH00358_96_AAFCFJGM5/kr19/intracellular_mutants_24h_a_2_S10_R2_001.fastq.gz -output intracellular_mutants_24h_run2 -primer AGCTTCAGGGTTGAGATGTGTATAAGAGACAG -mismatches 1 -bwa-alg mem mv tpp.cfg intracellular_mutants_24h_tpp_run2.cfg # "=SUM(B2:B130)" 20619934; "=SUM(C2:C130)" 1402849 # command: python /home/jhuang/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref merged_genome.fasta -reads1 ./240606_VH00358_96_AAFCFJGM5/kr19/intracellular_mutants_24h_a_2_S10_R1_001.fastq.gz -reads2 ./240606_VH00358_96_AAFCFJGM5/kr19/intracellular_mutants_24h_a_2_S10_R2_001.fastq.gz -output intracellular_mutants_24h_run2 -primer AGCTTCAGGGTTGAGATGTGTATAAGAGACAG -mismatches 1 -bwa-alg mem # transposon type: Tn5 # protocol type: Tn5 # bwa flags: # read1: ./240606_VH00358_96_AAFCFJGM5/kr19/intracellular_mutants_24h_a_2_S10_R1_001.fastq # read2: ./240606_VH00358_96_AAFCFJGM5/kr19/intracellular_mutants_24h_a_2_S10_R2_001.fastq # ref_genome: merged_genome.fasta # replicon_ids: # total_reads (or read pairs): 51244639 # truncated_reads 0 (genomic inserts shorter than the read length; ADAP2 appears in read1) # trimmed_reads (reads with valid Tn prefix, and insert size>20bp): 23204461 * # reads1_mapped: 20987078 # reads2_mapped: 20915836 # mapped_reads (both R1 and R2 map into genome, and R2 has a proper barcode): 20627374 * (since barcode is deleted, that means only this filters only the records in which the R2 not containing bacterial genome!) # read_count (TA sites only, for Himar1): 20627374 # template_count: 1405508 # template_ratio (reads per template): 14.68 # TA_sites: 4537463 # TAs_hit: 93859 # density: 0.021 # max_count (among templates): 285 # max_site (coordinate): 31693 # NZ_mean (among templates): 15.0 # FR_corr (Fwd templates vs. Rev templates): 0.003 # BC_corr (reads vs. templates, summed over both strands): 0.919 # Break-down of total reads (51244639): # 28040178 reads (54.7%) lack the expected Tn prefix # Break-down of trimmed reads with valid Tn prefix (23204461): # primer_matches: 0 reads (0.0%) contain CTAGAGGGCCCAATTCGCCCTATAGTGAGT (Himar1) # vector_matches: 0 reads (0.0%) contain CTAGACCGTCCAGTCTGGCAGGCCGGAAAC (phiMycoMarT7) # adapter_matches: 0 reads (0.0%) contain GATCGGAAGAGCACACGTCTGAACTCCAGTCAC (Illumina/TruSeq index) # misprimed_reads: 0 reads (0.0%) contain Himar1 prefix but don't end in TGTTA # read_length: 130 bp # mean_R1_genomic_length: 75.6 bp # mean_R2_genomic_length: 88.5 bp ./initial_mutants.tn_stats # Break-down of total reads (49821406): # 29481783 reads (59.2%) lack the expected Tn prefix # Break-down of trimmed reads with valid Tn prefix (20339623): --> #16,228,513 and 2,454,346 ./LB_culture.tn_stats # Break-down of total reads (43486192): # 20855173 reads (48.0%) lack the expected Tn prefix # Break-down of trimmed reads with valid Tn prefix (22631019): --> #19,735,541 and 3,266,320 ./growthout_control_24h.tn_stats # Break-down of total reads (70663823): # 43886543 reads (62.1%) lack the expected Tn prefix # Break-down of trimmed reads with valid Tn prefix (26777280): ./extracellular_mutants_24h.tn_stats # Break-down of total reads (47473664): # 38115004 reads (80.3%) lack the expected Tn prefix # Break-down of trimmed reads with valid Tn prefix (9358660): ./intracellular_mutants_24h.tn_stats # Break-down of total reads (51244639): # 28040178 reads (54.7%) lack the expected Tn prefix # Break-down of trimmed reads with valid Tn prefix (23204461): #grep "AGCTTCAGGGTTGAGATGTGTATAAGAGACAG" intracellular_mutants_24h_a_2_S10_R1_001.fastq | wc -l #28404198 #grep "AGCTTCAGGGTTGAGATGTGTATAAGAGACAG" intracellular_mutants_24h_a_2_S10_R2_001.fastq | wc -l #29 #AGCTTCAGGGTTGAGATGTGTATAAGAGACAG #NOTE_IMPORTANT: explain that some multiple mapped reads have to been deleted for the down-stream analysis! -
run Transposon Position Profiling (TPP) on the complete genome CP009367
#for initial_mutants python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref CP009367.fasta -reads1 240606_VH00358_96_AAFCFJGM5/kr11/initial_mutants_a_2_S6_R1_001.fastq.gz -reads2 240606_VH00358_96_AAFCFJGM5/kr11/initial_mutants_a_2_S6_R2_001.fastq.gz -output initial_mutants_run3 -primer AGCTTCAGGGTTGAGATGTGTATAAGAGACAG -mismatches 1 -bwa-alg mem mv tpp.cfg initial_mutants_tpp_run3.cfg #for LB_culture python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref CP009367.fasta -reads1 ./240606_VH00358_96_AAFCFJGM5/kr13/LB_culture_a_2_S7_R1_001.fastq.gz -reads2 ./240606_VH00358_96_AAFCFJGM5/kr13/LB_culture_a_2_S7_R2_001.fastq.gz -output LB_culture_run3 -primer AGCTTCAGGGTTGAGATGTGTATAAGAGACAG -mismatches 1 -bwa-alg mem mv tpp.cfg LB_culture_tpp_run3.cfg #for growthout_control_24h python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref CP009367.fasta -reads1 ./240606_VH00358_96_AAFCFJGM5/kr15/growthout_control_24h_a_2_S8_R1_001.fastq.gz -reads2 ./240606_VH00358_96_AAFCFJGM5/kr15/growthout_control_24h_a_2_S8_R2_001.fastq.gz -output growthout_control_24h_run3 -primer AGCTTCAGGGTTGAGATGTGTATAAGAGACAG -mismatches 1 -bwa-alg mem mv tpp.cfg growthout_control_24h_tpp_run3.cfg #for extracellular_mutants_24h python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref CP009367.fasta -reads1 ./240606_VH00358_96_AAFCFJGM5/kr17/extracellular_mutants_24h_a_2_S9_R1_001.fastq.gz -reads2 ./240606_VH00358_96_AAFCFJGM5/kr17/extracellular_mutants_24h_a_2_S9_R2_001.fastq.gz -output extracellular_mutants_24h_run3 -primer AGCTTCAGGGTTGAGATGTGTATAAGAGACAG -mismatches 1 -bwa-alg mem mv tpp.cfg extracellular_mutants_24h_tpp_run3.cfg #for intracellular_mutants_24h python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref CP009367.fasta -reads1 ./240606_VH00358_96_AAFCFJGM5/kr19/intracellular_mutants_24h_a_2_S10_R1_001.fastq.gz -reads2 ./240606_VH00358_96_AAFCFJGM5/kr19/intracellular_mutants_24h_a_2_S10_R2_001.fastq.gz -output intracellular_mutants_24h_run3 -primer AGCTTCAGGGTTGAGATGTGTATAAGAGACAG -mismatches 1 -bwa-alg mem mv tpp.cfg intracellular_mutants_24h_tpp_run3.cfg #NOTE that for some unknown reasons, intracellular_mutants_24h_run3.wig is always empty. Copy it for backup. cp intracellular_mutants_24h_run3.wig intracellular_mutants_24h_run3_backup.wig # "=SUM(B2:B130)" 20619934; "=SUM(C2:C130)" 1402849 ./initial_mutants_run3.tn_stats # Break-down of total reads (49821406): # 29481783 reads (59.2%) lack the expected Tn prefix # Break-down of trimmed reads with valid Tn prefix (20339623): ./LB_culture_run3.tn_stats # Break-down of total reads (43486192): # 20855173 reads (48.0%) lack the expected Tn prefix # Break-down of trimmed reads with valid Tn prefix (22631019): ./growthout_control_24h_run3.tn_stats # Break-down of total reads (70663823): # 43886543 reads (62.1%) lack the expected Tn prefix # Break-down of trimmed reads with valid Tn prefix (26777280): ./extracellular_mutants_24h_run3.tn_stats # Break-down of total reads (47473664): # 38115004 reads (80.3%) lack the expected Tn prefix # Break-down of trimmed reads with valid Tn prefix (9358660): ./intracellular_mutants_24h_run3.tn_stats # Break-down of total reads (51244639): # 28040178 reads (54.7%) lack the expected Tn prefix # Break-down of trimmed reads with valid Tn prefix (23204461): -
Prepare the wig files on merged_genome for transit running (it is not necessary for run3)
#change all wigs title to WA314 #./initial_mutants_run2.wig #./LB_culture_run2.wig #./growthout_control_24h_run2.wig #./intracellular_mutants_24h_run2.wig #./extracellular_mutants_24h_run2.wig sed -i 's/chrom=merged_genome/chrom=WA314/g' *_run2.wig -
Run Transit on merged_genome
#-- convert gbk or gff3 to prot_table -- #python3 ~/Scripts/gbk_to_prottable.py merged_genome.gbk merged_genome.prot_table python3 /home/jhuang/.local/bin/transit convert gff_to_prot_table CP009367.gff3 CP009367.prot_table #transit export #https://transit.readthedocs.io/en/latest/transit_normalization_tutorial.html #-1- transit export combined_wig <comma-separated .wig files> <annotation .prot_table> <output file> -n TTR transit export combined_wig initial_mutants_run3.wig,LB_culture_run3.wig,growthout_control_24h_run3.wig,intracellular_mutants_24h_run3.wig,extracellular_mutants_24h_run3.wig CP009367.prot_table combined.wig -n nonorm transit export combined_wig initial_mutants_run3.wig,LB_culture_run3.wig,growthout_control_24h_run3.wig,intracellular_mutants_24h_run3.wig,extracellular_mutants_24h_run3.wig CP009367.prot_table combined_normalized.wig -n TTR #-2- transit export igv initial_mutants_run3.wig,LB_culture_run3.wig,growthout_control_24h_run3.wig,intracellular_mutants_24h_run3.wig,extracellular_mutants_24h_run3.wig CP009367.prot_table combined.igv -n nonorm #[TO_SEND!] transit export igv initial_mutants_run3.wig,LB_culture_run3.wig,growthout_control_24h_run3.wig,intracellular_mutants_24h_run3.wig,extracellular_mutants_24h_run3.wig CP009367.prot_table combined_normalized.igv -n TTR #[TO_SEND!] #Open the two files in IGV, firstly import CP009367.gb, then import combined.igv or combined_normalized.igv. #-3- transit export mean_counts -c combined.wig combined.mean_counts #ARGS=['combined_on_CP009367.mean_counts'] #KWARGS={'c': 'combined_on_CP009367.wig'} #Error: list index out of range #python /home/jhuang/.local/bin/transit export mean_counts <comma-separated .wig files>|<combined_wig> <annotation .prot_table> <output file> [-c] #note: append -c if inputing a combined_wig file #https://training.galaxyproject.org/training-material/topics/genome-annotation/tutorials/tnseq/tutorial.html#compare-the-essential-genes-between-two-conditions # -- detect essential genes (DnaA is essential, which is why there are no insertion counts in the first few TA sites) #https://transit.readthedocs.io/en/v3.2.5/method_ttnfitness.html #对于 transit gumbel 分析,通常期望输入的是已经规范化(normalized)的 .wig 文件。规范化的数据可以减少由于不同实验条件或测序深度导致的偏差,使得后续的统计分析更加可靠 #-- Normalization -- #https://transit.readthedocs.io/en/latest/method_normalization.html for sample in initial_mutants LB_culture growthout_control_24h intracellular_mutants_24h extracellular_mutants_24h; do transit normalize ${sample}_run3.wig ${sample}_run3_normalized.wig -n TTR #betageom done #https://transit.readthedocs.io/en/latest/method_tnseq_stats.html #pre-normalization for sample in initial_mutants LB_culture growthout_control_24h intracellular_mutants_24h extracellular_mutants_24h; do transit tnseq_stats ${sample}_run3.wig done #post-normalization for sample in initial_mutants LB_culture growthout_control_24h intracellular_mutants_24h extracellular_mutants_24h; do transit tnseq_stats ${sample}_run3_normalized.wig done #File --> Export --> combined_wig, or IGV, or Mean_Gene_Counts. # --> Convert --> ...... [1] #View --> Scatter_Plot (only two datasets are allowed) # --> Track_View # --> Quality_Control #Analysis --> Himar1_Methods * gumbel * resampling * hmm * example * binomial * griffin * randproduct * utest * gi * normalize * tnseq_stats * corrplot * heatmap * ttnfitness --> Tn5_Methods * resampling: Resampling test of conditional essentiality between two conditions transit resampling -c combined.wig samples_run3.metadata LB_culture intracellular_mutants_24h CP009367.prot_table resampling_results_test.txt -s 10000 -n TTR -h -a -l -winz * utest: Mann-Whitney U-test of conditional essentiality between two conditions. This is a method for comparing datasets from a TnSeq library evaluated in two different conditions, analogous to resampling. transit utest LB_culture_run3.wig intracellular_mutants_24h_run3.wig CP009367.prot_table utest_out -n TTR -l #-l := Perform LOESS Correction; Helps remove possible genomic position bias. Default: Turned Off. * ZINB (command line only, If you want to compare more than two conditions, see ZINB.): The ZINB (Zero-Inflated Negative Binomial) method is used to determine which genes exhibit statistically significant variability across multiple conditions, in either the magnitude of insertion counts or local saturation, agnostically (in any one condition compared to the others). Like ANOVA, the ZINB method takes a combined_wig file (which combines multiple datasets in one file) and a samples_metadata file (which describes which samples/replicates belong to which experimental conditions). transit zinb combined.wig samples_run3.metadata CP009367.prot_table zinb_out -n TTR --condition Condition --include-conditions LB_culture,intracellular_mutants_24h #grep "not analyzed" zinb_out | wc -l #WARNING: Could run successful, but 4097 records are not analyzed! #TODO: R is called by Transit for certain commands, such as ZINB, corrplot, and heatmap. #install R (tested on v3.5.2) #R packages: MASS, pscl, corrplot, gplots (run install.packages(c("MASS", "pscl", "corrplot", "gplots")) install.packages("remotes") remotes::install_version("MASS", version = "7.3-60") #Python packages (for python3): rpy2 (v>=3.0) (run “pip3 install rpy2” on command line) ** ANOVA (command line only): The Anova (Analysis of variance) method is used to determine which genes exhibit statistically significant variability of insertion counts across multiple conditions. Unlike other methods which take a comma-separated list of wig files as input, the method takes a combined_wig file (which combined multiple datasets in one file) and a samples_metadata file (which describes which samples/replicates belong to which experimental conditions). transit anova combined.wig samples_run3.metadata CP009367.prot_table anova_out -n TTR --include-conditions LB_culture,intracellular_mutants_24h --ref LB_culture -PC 5 -alpha 1000 -winz #--ref <cond> := which condition(s) to use as a reference for calculating LFCs (comma-separated if multiple conditions) transit anova combined.wig samples_run3.metadata CP009367.prot_table anova_5samples_ref_LB_culture_out -n TTR --include-conditions initial_mutants,LB_culture,growthout_control_24h,intracellular_mutants_24h,extracellular_mutants_24h --ref LB_culture -PC 5 -alpha 1000 -winz #TODO_MERGE: merge combined.wig and combined_normalized.wig to Excel-file as the read_counts based on the gene! #Rv Gene TAs Mean_initial_mutants Mean_LB_culture Mean_growthout_control_24h Mean_intracellular_mutants_24h Mean_extracellular_mutants_24h LFC_initial_mutants LFC_LB_culture LFC_growthout_control_24h LFC_intracellular_mutants_24h LFC_extracellular_mutants_24h Fstat Pval #Padj status # Orf Gene ID. #Name Name of the gene. #TAs Number of TA sites in Gene #Means… Mean readcounts for each condition #LFCs… Log-fold-changes of counts in each condition vs mean across all conditions #MSR Mean-squared residual #MSE+alpha Mean-squared error, plus moderation value #p-value P-value calculated by the Anova test. #p-adj Adjusted p-value controlling for the FDR (Benjamini-Hochberg) #status Debug information (If any) transit example initial_mutants_run3.wig CP009367.prot_table initial_mutants_mean_read-counts_per_gene.txt #TODO: MERGE anovo+example together, delete all headers of the results, save as the Excel-file! # ---- commands for one sample ---- ** normalize: Normalization method: python transit.py norm glycerol_H37Rv_rep1.wig,glycerol_H37Rv_rep2.wig H37Rv.prot_table glycerol_TTR.txt -n TTR - TTR: Trimmed Total Reads (TTR), normalized by the total read-counts (like totreads), but trims top and bottom 5% of read-counts. This is the recommended normalization method for most cases, as it has the benefit of compensating for differences in saturation (which is especially important for resampling). - nzmean: Normalizes datasets to have the same mean over the non-zero sites. - totreads: Normalizes datasets by total read-counts, and scales them to have the same mean over all counts. - zinfnb: Fits a zero-inflated negative binomial model, and then divides read-counts by the mean. The zero-inflated negative binomial model will treat some empty sites as belonging to the “true” negative binomial distribution responsible for read-counts while treating the others as “essential” (and thus not influencing its parameters). - quantile: Normalizes datasets using the quantile normalization method described by Bolstad et al. (2003). In this normalization procedure, datasets are sorted, an empirical distribution is estimated as the mean across the sorted datasets at each site, and then the original (unsorted) datasets are assigned values from the empirical distribution based on their quantiles. This actually doesn’t work well on TnSeq data if a large fraction of TA sites have counts of 0 (ties). - betageom: Normalizes the datasets to fit an “ideal” Geometric distribution with a variable probability parameter p. Specially useful for datasets that contain a large skew. - nonorm: No normalization is performed. ** tnseq_stats: Statistical Metrics for TnSeq datasets #Typically a skew < 50 is desired #total_cts Sum of total read-counts in the sample. Indicates how much sequencing material was obtained. Typically >1M reads is desired for Himar1 datasets. transit tnseq_stats -c combined.wig -o tnseq_stats * example: Example method that calculates mean read-counts per gene. * transit example initial_mutants_run3.wig,LB_culture_run3.wig,growthout_control_24h_run3.wig,intracellular_mutants_24h_run3.wig,extracellular_mutants_24h_run3.wig CP009367.prot_table mean_read-counts_per_gene.txt * rankproduct: Rank product test for determining conditional essentiality. transit rankproduct LB_culture_run2.wig intracellular_mutants_24h_run2.wig merged_genome.prot_table rankproduct_out #-s 100 -n TTR -h -a -l transit rankproduct LB_culture_run3.wig intracellular_mutants_24h_run3.wig CP009367.prot_table rankproduct_out #ERROR! #Tn5Gaps method: https://training.galaxyproject.org/training-material/topics/genome-annotation/tutorials/tnseq/tutorial.html#predict-the-essentiality-of-genes ** tn5gaps: It is based on a Gumbel analysis method Griffin et al. 2011 and adapted to Tn5 transposon specificity. The main difference comes from the fact that Tn5 transposon can insert everywhere, thus creating libraries with lower insertion rates. #transit tn5gaps initial_mutants_run2.wig merged_genome.prot_table initial_mutants_tn5gaps_out #-m 2 -r Sum -iN 5 -iC 5; #transit tn5gaps initial_mutants_run3.wig CP009367.prot_table initial_mutants_tn5gaps_out for sample in initial_mutants LB_culture growthout_control_24h intracellular_mutants_24h extracellular_mutants_24h; do transit tn5gaps ${sample}_run3_normalized.wig CP009367.prot_table ${sample}_tn5gaps_trimmed.dat -m 2 -r Sum -iN 5 -iC 5; done #grep "Essential" initial_mutants_tn5gaps_trimmed.dat | wc -l #218 #grep "Non-essential" initial_mutants_tn5gaps_trimmed.dat | wc -l #3817 ~/Tools/csv2xls-0.4/csv_to_xls.py initial_mutants_tn5gaps_trimmed.dat LB_culture_tn5gaps_trimmed.dat \ growthout_control_24h_tn5gaps_trimmed.dat intracellular_mutants_24h_tn5gaps_trimmed.dat extracellular_mutants_24h_tn5gaps_trimmed.dat -d$'\t' -o Tn5Gaps.xls; # -- draw graphics to explain the r, ovr and lenovr based on the information below -- #Orf Name Desc k n r ovr lenovr pval padj call CH47_1 - 4Fe-4S dicluster domain protein 12 925 150 150 150 1.00000 1.00000 Non-essential CH47_10 hypE hydrogenase expression/formation protein HypE 8 871 349 349 349 0.99997 1.00000 Non-essential CH47_100 - hypothetical protein 21 150 26 26 26 1.00000 1.00000 Non-essential CH47_1000 - hypothetical protein 6 690 415 418 486 0.23409 1.00000 Non-essential k: Number of Transposon Insertions Observed within the ORF. n: Total Number of TA dinucleotides within the ORF. r: Length of the Maximum Run of Non-Insertions observed. #TODO1_DEL: ovr: The number of nucleotides in the overlap with the longest run partially covering the gene. lenovr: The length of the above run with the largest overlap with the gene. pval: P-value calculated by the permutation test. padj: Adjusted p-value controlling for the FDR (Benjamini-Hochberg). call: Essentiality call for the gene. Depends on FDR corrected thresholds. Essential or Non-Essential. r (Run of Non-Insertions): This value represents the length of the longest continuous region within an ORF (open reading frame) where no transposon insertions are observed. Graphically, this could be shown as a long unbroken line or bar within a longer gene representation, highlighting the absence of marks (insertions). ovrovr (Overlap with Run): This is the number of nucleotides that overlap with the longest run, which might partially cover the gene. It is not the total length of the run, but how much of it overlaps with the gene. In a graphic, this could be illustrated by overlapping two segments: one for the gene and another for the run, with the overlapping part distinctly colored or shaded. lenovrlenovr (Length of Overlap with Run): This measures the full length of the run that has the largest overlap with the gene. Visually, this could be depicted as a separate longer line or bar that extends beyond the gene boundaries but is highlighted where it overlaps with the gene. * #WARNING: Since gumbel_out cannot be generated, the ttnfitness can be generated! * TTN-Fitness (TTNFitness method that calculates mean read-counts per gene): Typically with individual TnSeq datasets, Gumbel and HMM are the methods used for evaluating essentiality. - Gumbel distinguishes between ES (essential) from NE (non-essential). - HMM adds the GD (growth-defect; suppressed counts; mutant has reduced fitness) and GA (growth advantage; inflated counts; mutant has selective advantage) categories. - Quantifying the magnitude of the fitness defect is risky because the counts at individual TA sites can be noisy. Sometimes the counts at a TA site in a gene can span a wide range of very low to very high counts. The TTN-Fitness gives a more fine-grained analysis of the degree of fitness effect by taking into account the insertion preferences of the Himar1 transposon. - These insertion preferences are influenced by the nucleotide context of each TA site. The TTN-Fitness method uses a statistical model based on surrounding nucleotides to estimate the insertion bias of each site. Then, it corrects for this to compute an overall fitness level as a Fitness Ratio, where the ratio is 0 for ES genes, 1 for typical NE genes, between 0 and 1 for GD genes and above 1 for GA genes. #transit ttnfitness <comma-separated .wig files> <annotation .prot_table> <genome .fna> <gumbel output file> <output1 file> <output2 file> transit ttnfitness initial_mutants_run3_normalized.wig,LB_culture_run3_normalized.wig,growthout_control_24h_run3_normalized.wig,intracellular_mutants_24h_run3_normalized.wig,extracellular_mutants_24h_run3_normalized.wig CP009367.prot_table CP009367.fa gumbel_out ttnfitness_out1 ttnfitness_out2 > ttnfitness_log #ERROR - gumbel output file:* The Gumbel method must be run first on the dataset.The output of the Gumbel method is provided as an input to this method. ES (essential by Gumbel) and EB (essential by Binomial) is calculated in the TTN-Fitness method via this files * Genetic Interactions: The genetic interactions (GI) method is a comparative analysis used used to determine genetic interactions. It is a Bayesian method that estimates the distribution of log fold-changes (logFC) in two strain backgrounds under different conditions, and identifies significantly large changes in enrichment (delta_logFC) to identify those genes that imply a genetic interaction. * Pathway Enrichment Analysis: Pathway Enrichment Analysis provides a method to identify enrichment of functionally-related genes among those that are conditionally essential (i.e. significantly more or less essential between two conditions). transit pathway_enrichment <resampling_file> <associations> <pathways> <output_file> [-M <FET|GSEA|GO>] [- ... ** corrplot (Correlation among TnSeq datasets, command line only): A useful tool when evaluating the quality of a collection of TnSeq datasets is to make a correlation plot of the mean insertion counts (averaged at the gene-level) among samples. #INCOMPLETE cut -f1-6 combined.wig > combined_.wig transit corrplot combined_.wig corrplot.png #INCOMPLETE cut -f1-6 combined_normalized.wig > combined_normalized_.wig #transit corrplot combined_normalized_.wig corrplot_normalized.png transit corrplot anova_5samples_ref_LB_culture_out corrplot_anova.png -anova ** heatmap (Heatmap among Conditions, command line only): The output of ANOVA or ZINB can be used to generate a heatmap that simultaneously clusters the significant genes and clusters the conditions, which is especially useful for shedding light on the relationships among the conditions apparent in the data. transit heatmap anova_5samples_ref_LB_culture_out heatmap.png -anova -qval 0.05 -low_mean_filter 3 #heatmap based on 74 genes transit heatmap anova_5samples_ref_LB_culture_out heatmap.png -anova -qval 0.1 -low_mean_filter 3 -
Reports
#1. For gene_based reports: example (mean_read-counts_per_gene.txt) + ANOVA (anova_5samples_ref_LB_culture_out) #TAs: Number of TA sites in Gene #mean: Average read-count, including empty sites. #nzmean: Average read-count, excluding empty sites. Orf: Gene ID Name: Name of the gene. Desc Gene description. k: Number of Transposon Insertions Observed within the ORF. n: Total Number of TA sites within the ORF Normalized_initial_mutants: Mean read counts for the condition initial_mutants (normalized with TTR) Normalized_LB_culture: Mean read counts for the condition LB_culture (normalized with TTR) Normalized_growthout_control_24h: Mean read counts for the condition growthout_control_24h (normalized with TTR) Normalized_intracellular_mutants_24h: Mean read counts for the condition intracellular_mutants_24h (normalized with TTR) Normalized_extracellular_mutants_24h: Mean read counts for the condition extracellular_mutants_24h (normalized with TTR) #check both f1 are the same; sort -t'_' -k2,2n mean_read-counts_per_gene.txt -o mean_read-counts_per_gene_sorted.txt sort -t'_' -k2,2n anova_5samples_ref_LB_culture_out -o anova_5samples_ref_LB_culture_out_sorted #DELETE all header lines except for keeping one in sorted files. cut -f1 mean_read-counts_per_gene_sorted.txt > f1_1 cut -f1 anova_5samples_ref_LB_culture_out_sorted > f1_2 diff f1_1 f1_2 #Orf Name Desc k n mean nzmean cut -f1-5 mean_read-counts_per_gene_sorted.txt > f1_5; #delete the columns mean and nzmean. #Rv Gene TAs Normalized_initial_mutants Normalized_LB_culture Normalized_growthout_control_24h Normalized_intracellular_mutants_24h Normalized_extracellular_mutants_24h cut -f4-8 anova_5samples_ref_LB_culture_out_sorted > f4_8; paste f1_5 f4_8 > overview_gene_based.txt #~/Tools/csv2xls-0.4/csv_to_xls.py overview_gene_based.txt -d$'\t' -o overview_gene_based.xls; #2. For essentiall gene report: transit tn5gaps #Tn5Gaps method: https://training.galaxyproject.org/training-material/topics/genome-annotation/tutorials/tnseq/tutorial.html#predict-the-essentiality-of-genes #dinucleotides ORF Gene ID. Name Name of the gene. Desc Gene description. k Number of Transposon Insertions Observed within the ORF. n Total Number of TA sites within the ORF. r Length of the Maximum Run of Non-Insertions observed. pval P-value calculated by the permutation test. padj Adjusted p-value controlling for the FDR (Benjamini-Hochberg). call Essentiality call for the gene. Depends on FDR corrected thresholds. Essential or Non-Essential. #DELETE the headers; DEL the columns ovr and lenovr; merge tn5gaps also the gene-based tables in the Tn5Gaps.xls. #3. Only choose ANOVA for DEG reports, since it can be drawn + are consistent with the gene-based view! #Orf: Gene ID #Name: Name of the gene #TAs: Number of TA sites in the gene #Means…: Mean read counts for each condition #LFCs…: Log-fold changes of counts in each condition vs. the mean across all conditions #p-value: P-value calculated by the ANOVA test #p-adj: Adjusted p-value controlling for the FDR (False Discovery Rate, Benjamini-Hochberg method) #status: Debug information #~/Tools/csv2xls-0.4/csv_to_xls.py anova_out -d$'\t' -o anova_out.xls; # DELETE the headers and the columns MSR and MSE+alpha; SORT according to Padj. #4. 2 plots corrplot: A useful tool when evaluating the quality of a collection of TnSeq datasets is to make a correlation plot of the mean insertion counts (averaged at the gene-level) among samples. heatmap: The output of ANOVA can be used to generate a heatmap that simultaneously clusters the significant genes and clusters the conditions, which is especially useful for shedding light on the relationships among the conditions apparent in the data. #5. igv and genbank files for IGV opening #Usage: Open the two files in IGV, firstly import CP009367.gb, then import combined.igv or combined_normalized.igv. -
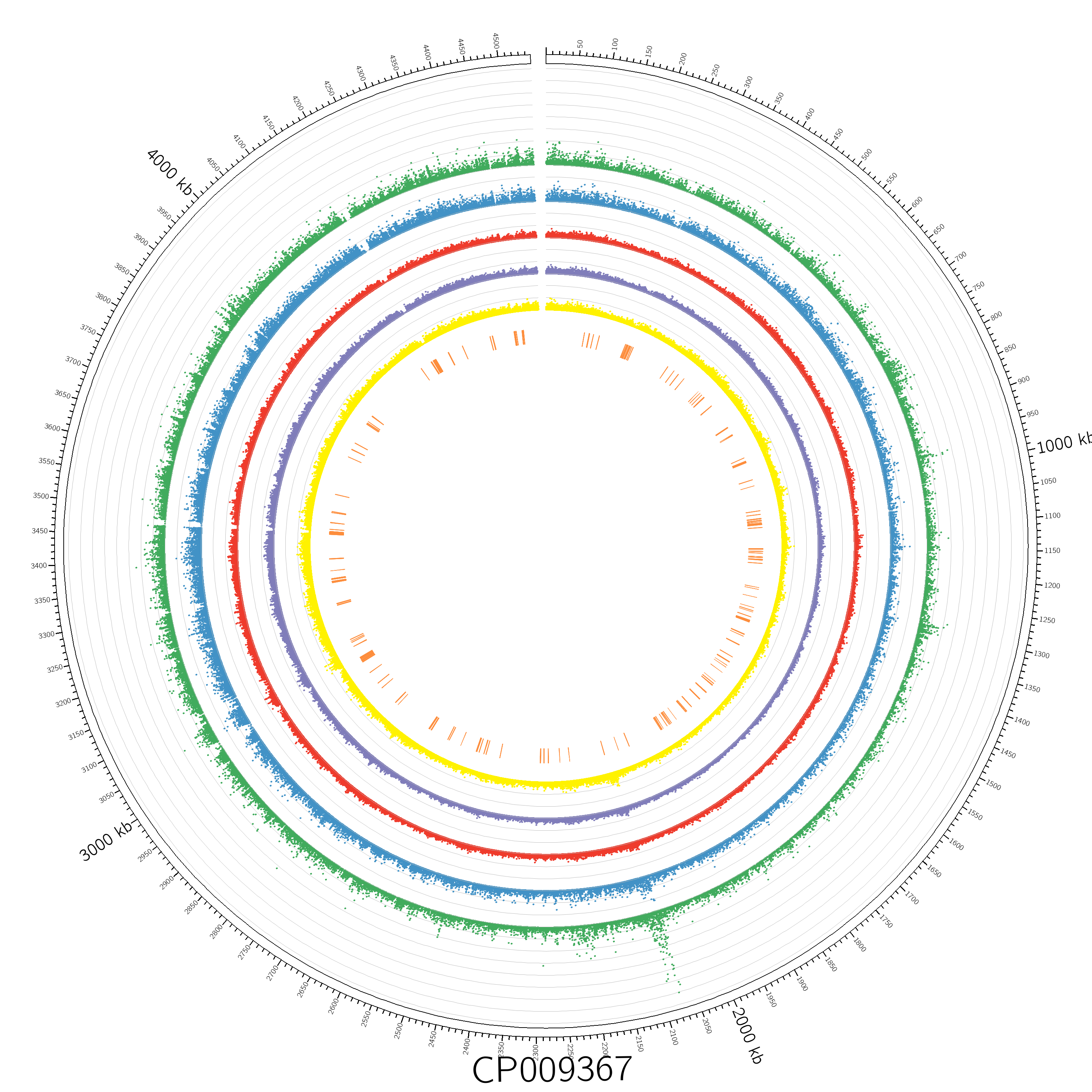
plot Figure 1. Overview of the Yersinia enterocolitica subsp. enterocolitica WA-314 Transposon Mutant Library This figure illustrates the distribution of transposon insertion sites across the genome. The outermost black circle represents the WA-314 genome in kilobase pairs (Kbp). The middle circles, which feature scatter points, indicate the normalized number of sequencing reads at each unique transposon insertion site, with each axis line representing an increment of 50,000 in data values. The circles are color-coded to represent different conditions: green for extracellular mutants, blue for intracellular mutants, red for growthout control, purple for LB culture, and yellow for initial mutants. The innermost orange circle highlights the locations of genes identified as essential.
#create karyotype.microbe.txt contains the following line chr - chr CP009367 0 4548749 black #Input file: combined_normalized.wig #Output file: initial_mutants.txt LB_culture.txt growthout_control.txt intracellular_mutants.txt extracellular_mutants.txt python3 generate_circos_input_files.py #Prepare CP009367.prot_table_essential from CP009367.prot_table and initial_mutants_tn5gaps_trimmed.dat grep "Essential" initial_mutants_tn5gaps_trimmed.dat > essential.ids cut -f1 essential.ids > essential.f1 #DELETE the header #replace \t with | in CP009367.prot_table_divided_with_ #replace \n with |" CP009367.prot_table_divided_with_ >> CP009367.prot_table_essential\ngrep "| in essential.f1 #replace | with \t in CP009367.prot_table_essential #format circos_data/merged_genome.txt to the following format #chr 98090 98972 1 #chr 99072 100149 1 #... circos -conf circos.conf -
generate_circos_input_files.py
import pandas as pd import os # Function to read the combined .wig file def read_combined_wig(file): with open(file, 'r') as f: lines = f.readlines() header_line = next(i for i, line in enumerate(lines) if line.startswith("#TA_coord")) + 1 data = pd.read_csv(file, sep="\t", skiprows=header_line, header=None) data.columns = ["TA_coord", "initial_mutants", "LB_culture", "growthout_control", "intracellular_mutants", "extracellular_mutants", "gene_id"] data["TA_coord"] = pd.to_numeric(data["TA_coord"]) return data # Read the .wig file wig_file = "combined_normalized.wig" plot_data = read_combined_wig(wig_file) # Melt the DataFrame for easier manipulation plot_data_long = plot_data.melt(id_vars=["TA_coord", "gene_id"], var_name="condition", value_name="value") # Filter out zeros for better visualization (optional) plot_data_long = plot_data_long[plot_data_long["value"] > 0] # Ensure the condition column is treated as a category plot_data_long["condition"] = plot_data_long["condition"].astype("category") # Function to save data for a specific condition def save_condition_data(data, condition, filename): condition_data = data[data["condition"] == condition].copy() condition_data["chr"] = "chr" condition_data["start"] = condition_data["TA_coord"] condition_data["end"] = condition_data["TA_coord"] + 1 condition_data = condition_data[["chr", "start", "end", "value"]] condition_data.to_csv(filename, sep="\t", header=False, index=False) # Create data directory if it doesn't exist if not os.path.exists("circos_data"): os.makedirs("circos_data") # Save data for each condition save_condition_data(plot_data_long, "initial_mutants", "circos_data/initial_mutants.txt") save_condition_data(plot_data_long, "LB_culture", "circos_data/LB_culture.txt") save_condition_data(plot_data_long, "growthout_control", "circos_data/growthout_control.txt") save_condition_data(plot_data_long, "intracellular_mutants", "circos_data/intracellular_mutants.txt") save_condition_data(plot_data_long, "extracellular_mutants", "circos_data/extracellular_mutants.txt") -
generate_gene_locations.py
import pandas as pd # Read the merged genome table prot_table = pd.read_csv("CP009367.prot_table_essential", sep="\t", header=None) # Print the column names to check for correctness print(prot_table.columns) # Create a new DataFrame with required columns for Circos circos_genes = pd.DataFrame() circos_genes["chr"] = "chr" # Chromosome name (constant for all entries) # Use correct column indices based on inspection circos_genes["start"] = prot_table.iloc[:, 1] # Second column for start coordinate circos_genes["end"] = prot_table.iloc[:, 2] # Third column for end coordinate circos_genes["value"] = 1 # Set a constant value for heatmap # Save the Circos gene location file circos_genes.to_csv("circos_data/merged_genome.txt", sep="\t", header=False, index=False) -
circos.conf
#circos -conf circos.conf <<include /etc/circos/colors_fonts_patterns.conf>> karyotype = circos_data/karyotype.microbe.txt chromosomes_units = 10000 chromosomes_display_default = yes <ideogram> show = yes <spacing> default = 0.01r break = 0.5r #8r # Adds a gap between the first and last position of the single chromosome </spacing> radius = 0.9r thickness = 25p fill = no stroke_thickness = 2 stroke_color = black show_bands = yes fill_bands = yes band_transparency = 0 show_label = yes label_font = default label_radius = 1.05r label_size = 75 label_parallel = yes orientation = 100 # Rotate the plot by 90 degrees </ideogram> #<<include ticks.conf>> show_ticks = yes show_tick_labels = yes show_grid = no grid_start = dims(ideogram,radius_inner)-0.5r grid_end = dims(ideogram,radius_inner) <ticks> skip_first_label = yes skip_last_label = no radius = dims(ideogram,radius_outer) tick_separation = 2p min_label_distance_to_edge = 0p label_separation = 5p label_offset = 5p label_size = 8p multiplier = 0.001 color = black thickness = 3p size = 20p <tick> size = 10p spacing = 1u color = black show_label = no label_size = 12p format = %.2f grid = no grid_color = lblue grid_thickness = 1p </tick> <tick> size = 15p spacing = 5u color = black show_label = yes label_size = 16p format = %s grid = yes grid_color = lgrey grid_thickness = 1p </tick> <tick> size = 18p spacing = 10u color = black show_label = yes label_size = 16p format = %s grid = yes grid_color = grey grid_thickness = 1p </tick> <tick> spacing = 100u color = black show_label = yes suffix = " kb" label_size = 36p format = %s grid = yes grid_color = dgrey grid_thickness = 1p </tick> </ticks> <image> <<include /etc/circos/image.conf>> </image> <colors> <<include /etc/circos/colors.conf>> </colors> <fonts> <<include /etc/circos/fonts.conf>> </fonts> # -- Scatter plot 1 -- <plots> <plot> type = scatter file = circos_data/extracellular_mutants.txt r1 = 0.99r r0 = 0.79r #python3 identify_min_max.py circos_data/initial_mutants.txt min = 0 max = 400000 glyph = circle glyph_size = 5 color = dgreen <axes> <axis> spacing = 50000 color = lgrey </axis> </axes> #<axes> #<axis> #spacing = 0.1r #color = lgrey #<ticks> #<tick> #spacing = 0.1 #size = 10p #thickness = 2p #color = lgrey #show_label = yes #label_size = 20p #label_offset = 5p #format = %0.1f #</tick> #</ticks> #</axis> #</axes> #<rules> #<rule> #condition = var(value) > 10000 #stroke_color = dred #fill_color = red #glyph = rectangle #glyph_size = 2 #</rule> #</rules> </plot> # -- Scatter plot 2 -- <plot> type = scatter file = circos_data/intracellular_mutants.txt r1 = 0.79r r0 = 0.715r min = 0 max = 150000 glyph = circle glyph_size = 5 color = dblue <axes> <axis> spacing = 50000 color = lgrey </axis> </axes> </plot> # -- Scatter plot 3 -- <plot> type = scatter file = circos_data/growthout_control.txt r1 = 0.715r r0 = 0.64r min = 0 max = 150000 glyph = circle glyph_size = 5 color = dred <axes> <axis> spacing = 50000 color = lgrey </axis> </axes> </plot> # -- Scatter plot 4 -- <plot> type = scatter file = circos_data/LB_culture.txt r1 = 0.64r r0 = 0.565r min = 0 max = 150000 glyph = circle glyph_size = 5 color = dpurple <axes> <axis> spacing = 50000 color = lgrey </axis> </axes> </plot> # -- Scatter plot 5 -- <plot> type = scatter file = circos_data/initial_mutants.txt r1 = 0.565r r0 = 0.49r min = 0 max = 150000 glyph = circle glyph_size = 5 color = dyellow <axes> <axis> spacing = 50000 color = lgrey </axis> </axes> </plot> # Gene Locations <plot> type = heatmap file = circos_data/merged_genome.txt r1 = 0.45r r0 = 0.42r color = orange </plot> #grep "Essential" tn5_gap_inituial_mutant.csv > essential_genes.txt #cut -f1 -d$'\t' essential_genes.txt > f1 #vim merged_genome.prot_table #" merged_genome.prot_table >> merged_genome.prot_table_essential \ngrep " #python3 generate_gene_locations.py #replace "\n\t" with "\nchr\t" </plots> <<include /etc/circos/housekeeping.conf>>
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Kraken2 Installation and Usage Guide
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Should the inputs for GSVA be normalized or raw?
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- Risks of Rebooting into Rescue Mode
- 足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Processing Data_Michelle_RNAseq_2025
Setup the environment for lumicks-pylake and C_Trap-Multimer-photontrack.ipynb
🧬 Cadmium Resistance Gene Analysis in Staphylococcus epidermidis HD46
Workflow for RNA-Binding Protein Enrichment and RNA Type Distribution Analysis (Ute’s Project)