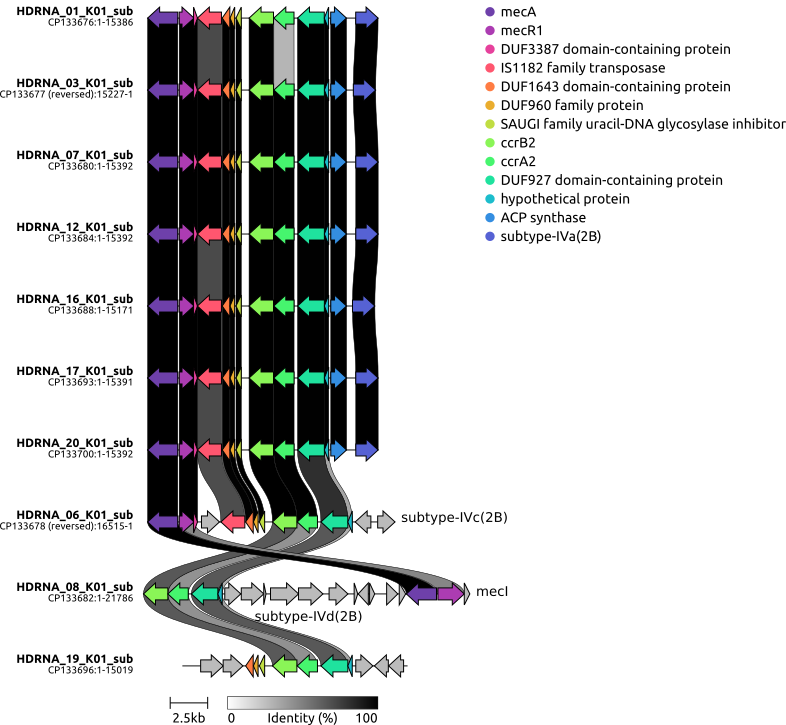
clinker usage
gene_x 0 like s 920 view s
Tags: pipeline
https://github.com/gamcil/clinker/
https://cagecat.bioinformatics.nl/
https://pegaso.microbiologia.ull.es/clinker-server.php
-
code of extract_subregion.py
import sys from Bio import SeqIO def extract_subregion(genbank_path, start, end, output_file): for record in SeqIO.parse(genbank_path, "genbank"): subregion = record[start:end] SeqIO.write(subregion, output_file, "genbank") if __name__ == "__main__": if len(sys.argv) < 5: print("Usage: python extract_subregion.py <GenBank file> <start> <end> <output file>") sys.exit(1) genbank_file = sys.argv[1] start = int(sys.argv[2]) end = int(sys.argv[3]) output_file = sys.argv[4] extract_subregion(genbank_file, start, end, output_file) -
commands
python3 extract_subregion.py HDRNA_01_K01_conservative_23197.current.gb 40434 55820 HDRNA_01_K01_sub.gb python3 extract_subregion.py HDRNA_03_K01_bold_bandage_26831.current.gb 2112517 2127744 HDRNA_03_K01_sub.gb python3 extract_subregion.py HDRNA_06_K01_conservative_27645.current_chr.gb 42053 58568 HDRNA_06_K01_sub.gb python3 extract_subregion.py HDRNA_07_K01_conservative_27169.current_chr.gb 37601 52993 HDRNA_07_K01_sub.gb python3 extract_subregion.py HDRNA_08_K01_conservative_32455.current_chr.gb 47084 68870 HDRNA_08_K01_sub.gb python3 extract_subregion.py HDRNA_12_K01_bold_37467.current_chr.gb 37586 52978 HDRNA_12_K01_sub.gb python3 extract_subregion.py HDRNA_16_K01_conservative_37834.current_chr.gb 37706 52877 HDRNA_16_K01_sub.gb python3 extract_subregion.py HDRNA_17_K01_conservative_37288.current_chr.gb 37118 52509 HDRNA_17_K01_sub.gb python3 extract_subregion.py HDRNA_19_K01_bold_37377.current_chr.gb 36267 51286 HDRNA_19_K01_sub.gb python3 extract_subregion.py HDRNA_20_K01_conservative_43457.current_chr.gb 37703 53095 HDRNA_20_K01_sub.gb clinker gbk_sub/*.gb -p plot.html --dont_set_origin -s session.json -o alignments.csv -dl "," -dc 4
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Kraken2 Installation and Usage Guide
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Should the inputs for GSVA be normalized or raw?
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- Risks of Rebooting into Rescue Mode
- 足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Processing Data_Michelle_RNAseq_2025
Setup the environment for lumicks-pylake and C_Trap-Multimer-photontrack.ipynb
🧬 Cadmium Resistance Gene Analysis in Staphylococcus epidermidis HD46
Workflow for RNA-Binding Protein Enrichment and RNA Type Distribution Analysis (Ute’s Project)