Analysis of SNPs, InDels, transposons, and IS elements in 5 A. baumannii strains
gene_x 0 like s 942 view s
Tags: variation
1, call variant calling using snippy
mkdir snippy_CP059040;
for sample in snippy_CP059040; do
cd ${sample};
# -- hard-copy --
#git clone https://github.com/huang/bacto
#mv bacto/* ./
#rm -rf bacto
# -- soft-link --
ln -s ~/Tools/bacto/db/ .;
ln -s ~/Tools/bacto/envs/ .;
ln -s ~/Tools/bacto/local/ .;
cp ~/Tools/bacto/Snakefile .;
cp ~/Tools/bacto/bacto-0.1.json .;
cp ~/Tools/bacto/cluster.json .;
cd ..;
done
mkdir raw_data; cd raw_data;
ln -s ../../01.RawData/Tig1/Tig1_1.fq.gz Tig1_R1.fastq.gz
ln -s ../../01.RawData/Tig1/Tig1_2.fq.gz Tig1_R2.fastq.gz
ln -s ../../01.RawData/Tig2/Tig2_1.fq.gz Tig2_R1.fastq.gz
ln -s ../../01.RawData/Tig2/Tig2_2.fq.gz Tig2_R2.fastq.gz
ln -s ../../01.RawData/W/W_1.fq.gz W_R1.fastq.gz
ln -s ../../01.RawData/W/W_2.fq.gz W_R2.fastq.gz
ln -s ../../01.RawData/Y/Y_1.fq.gz Y_R1.fastq.gz
ln -s ../../01.RawData/Y/Y_2.fq.gz Y_R2.fastq.gz
ln -s ../../01.RawData/△adeIJ/△adeIJ_1.fq.gz _adeIJ_R1.fastq.gz
ln -s ../../01.RawData/△adeIJ/△adeIJ_2.fq.gz _adeIJ_R2.fastq.gz
#download CP059040.gb from GenBank
mv ~/Downloads/sequence\(1\).gb db/CP059040.gb
#setting the following in bacto-0.1.json
"genus": "Acinetobacter",
"kingdom": "Bacteria",
"species": "baumannii",
"reference": "db/CP059040.gb"
conda activate bengal3_ac3
(bengal3_ac3) cd snippy_CP059040
(bengal3_ac3) /home/jhuang/miniconda3/envs/snakemake_4_3_1/bin/snakemake --printshellcmds
2, using spandx calling variants (almost the same results to the one from viral-ngs!)
mkdir ~/miniconda3/envs/spandx/share/snpeff-5.1-2/data/CP059040
cp CP059040.gb ~/miniconda3/envs/spandx/share/snpeff-5.1-2/data/CP059040/genes.gbk
vim ~/miniconda3/envs/spandx/share/snpeff-5.1-2/snpEff.config
/home/jhuang/miniconda3/envs/spandx/bin/snpEff build CP059040 #-d
~/Scripts/genbank2fasta.py CP059040.gb
mv CP059040.gb_converted.fna CP059040.fasta #rename "CP059040.1 xxxxx" to "CP059040" in the fasta-file
ln -s /home/jhuang/Tools/spandx/ spandx
(spandx) nextflow run spandx/main.nf --fastq "snippy_CP059040/trimmed/*_P_{1,2}.fastq" --ref CP059040.fasta --annotation --database CP059040 -resume
3, summarize all SNPs and Indels from the snippy result directory.
#Output: snippy_CP059040/snippy/summary_snps_indels.csv
# adapt the array isolates = ["Tig1", "Tig2", "Y", "W", "_adeIJ"]
python3 ~/Scripts/summarize_snippy_res.py snippy_CP059040/snippy
grep -v "None,,,,,,None,None" summary_snps_indels.csv > summary_snps_indels_.csv
grep -v "/" All_SNPs_indels_annotated.txt > All_SNPs_indels_annotated_.txt
4, merge the two files summary_snps_indels_.csv and All_SNPs_indels_annotated_.txt by merging the results from two variant calling methods snippy and spandx
cut -d$'\t' -f2 All_SNPs_indels_annotated_.txt > ../../id1
cut -d',' -f2 summary_snps_indels_.csv > ../../id2
diff id1 id2
5, (optional, since it should not happed) filter rows without change (between REF and the isolates) in snippy_CP059040/snippy/summary_snps_indels_.csv (94)
awk -F, '
NR == 1 {print; next} # Print the header line
{
ref = $3; # Reference base (assuming REF is in the 3rd column)
same = 1; # Flag to check if all bases are the same as reference
samples = ""; # Initialize variable to hold sample data
for (i = 6; i <= NF - 8; i++) { # Loop through all sample columns, adjusting for the number of fixed columns
samples = samples $i " "; # Collect sample data
if ($i != ref) {
same = 0;
}
}
if (!same) {
print $0; # Print the entire line if not all bases are the same as the reference
#print "Samples: " samples; # Print all sample data for checking
}
}
' merged_variants_CP133676.csv > merged_variants_CP133676_.csv
#Explanation:
# -F, sets the field separator to a comma.
# NR == 1 {print; next} prints the header line and skips to the next line.
# ref = $3 sets the reference base (assumed to be in the 3rd column).
# same = 1 initializes a flag to check if all sample bases are the same as the reference.
# samples = ""; initializes a string to collect sample data.
# for (i = 6; i <= NF - 8; i++) loops through the sample columns. This assumes the first 5 columns are fixed (CHROM, POS, REF, ALT, TYPE), and the last 8 columns are fixed (Effect, Impact, Functional_Class, Codon_change, Protein_and_nucleotide_change, Amino_Acid_Length, Gene_name, Biotype).
# samples = samples $i " "; collects sample data.
# if ($i != ref) { same = 0; } checks if any sample base is different from the reference base. If found, it sets same to 0.
# if (!same) { print $0; print "Samples: " samples; } prints the entire line and the sample data if not all sample bases are the same as the reference.
6, improve the header
sed -i '1s/_trimmed_P//g' merged_variants_CP059040.csv
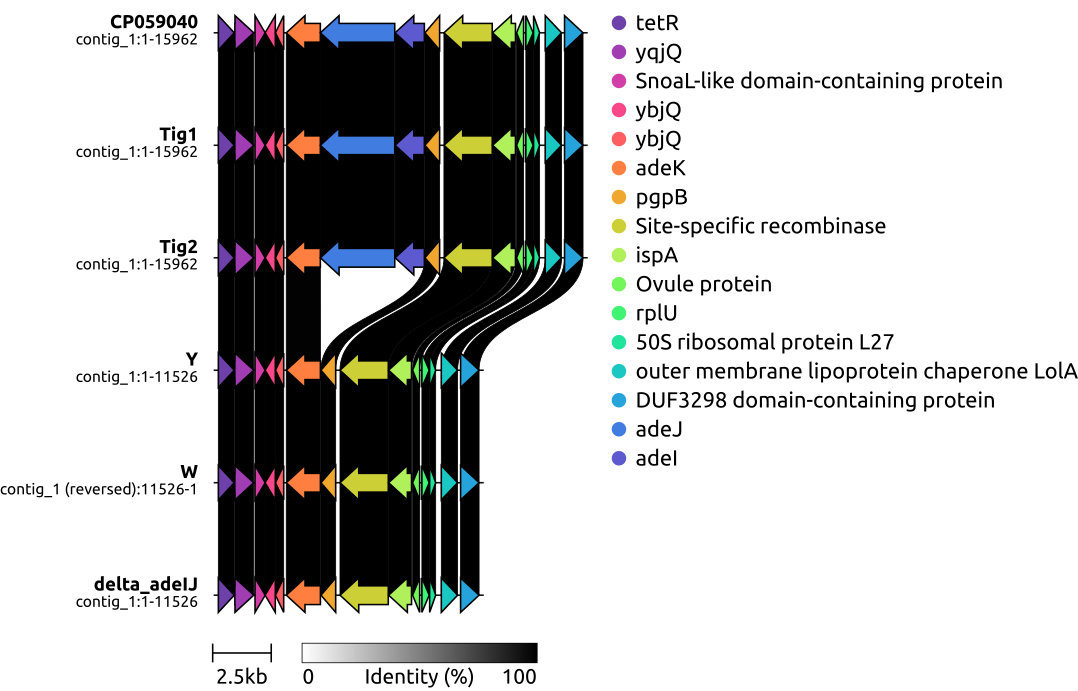
7, draw local genetic environments of △adeIJ
mkdir gbff_files
(bakta) bakta --db /mnt/nvme0n1p1/REFs/bakta_db snippy_CP059040/shovill/Tig1/contigs.fa --prefix Tig1
(bakta) bakta --db /mnt/nvme0n1p1/REFs/bakta_db snippy_CP059040/shovill/Tig2/contigs.fa --prefix Tig2
(bakta) bakta --db /mnt/nvme0n1p1/REFs/bakta_db snippy_CP059040/shovill/Y/contigs.fa --prefix Y
(bakta) bakta --db /mnt/nvme0n1p1/REFs/bakta_db snippy_CP059040/shovill/W/contigs.fa --prefix W
(bakta) bakta --db /mnt/nvme0n1p1/REFs/bakta_db snippy_CP059040/shovill/_adeIJ/contigs.fa --prefix _adeIJ
# -- find the gene positions in the gff3-file --
#tetR: 204019
#adeJ: complement(208370..211546)
#adeI: complement(211559..212809)
#DUF3298 domain-containing protein 218792..219580
#python3 ~/Scripts/extract_subregion.py Tig1.gbff 203819 219780 adeIJ_sub/Tig1.gbff
samtools faidx Tig1.fna
samtools faidx Tig1.fna contig_7:203819-219780 > Tig1_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db Tig1_sub.fna
mv Tig1_sub.gbff gbff_files/Tig1.gbff
samtools faidx Tig2.fna
samtools faidx Tig2.fna contig_7:206569-222530 > Tig2_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db Tig2_sub.fna
mv Tig2_sub.gbff gbff_files/Tig2.gbff
samtools faidx Y.fna
samtools faidx Y.fna contig_8:203819-215344 > Y_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db Y_sub.fna
mv Y_sub.gbff gbff_files/Y.gbff
samtools faidx W.fna
samtools faidx W.fna contig_7:37321-48846 > W_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db W_sub.fna
mv W_sub.gbff gbff_files/W.gbff
samtools faidx _adeIJ.fna
samtools faidx _adeIJ.fna contig_7:203819-215344 > _adeIJ_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db _adeIJ_sub.fna
mv _adeIJ_sub.gbff gbff_files/delta_adeIJ.gbff
makeblastdb -in ~/DATA/Data_Tam_variant_calling/snippy_CP059040/db/CP059040.gb_converted.fna -dbtype 'nucl' -out CP059040.gb_converted.fna.db
blastn -db CP059040.gb_converted.fna.db -query Tig1_sub.fna -out adeKJI_on_CP059040.blastn -evalue 1e-10 -num_threads 15 -outfmt 6
samtools faidx ~/DATA/Data_Tam_variant_calling/snippy_CP059040/db/CP059040.gb_converted.fna
samtools faidx ~/DATA/Data_Tam_variant_calling/snippy_CP059040/db/CP059040.gb_converted.fna CP059040:732682-748643 > CP059040_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db CP059040_sub.fna
mv CP059040_sub.gbff gbff_files/CP059040.gbff
cd gbff_files
rm *.json
clinker *.gbff -p plot.html --dont_set_origin -s session.json -o alignments.csv -dl "," -dc 4
#~/Tools/csv2xls-0.4/csv_to_xls.py Tig1_.gff3 Tig2_.gff3 Y_.gff3 W_.gff3 _adeIJ_.gff3 -d$'\t' -o gff3.xls
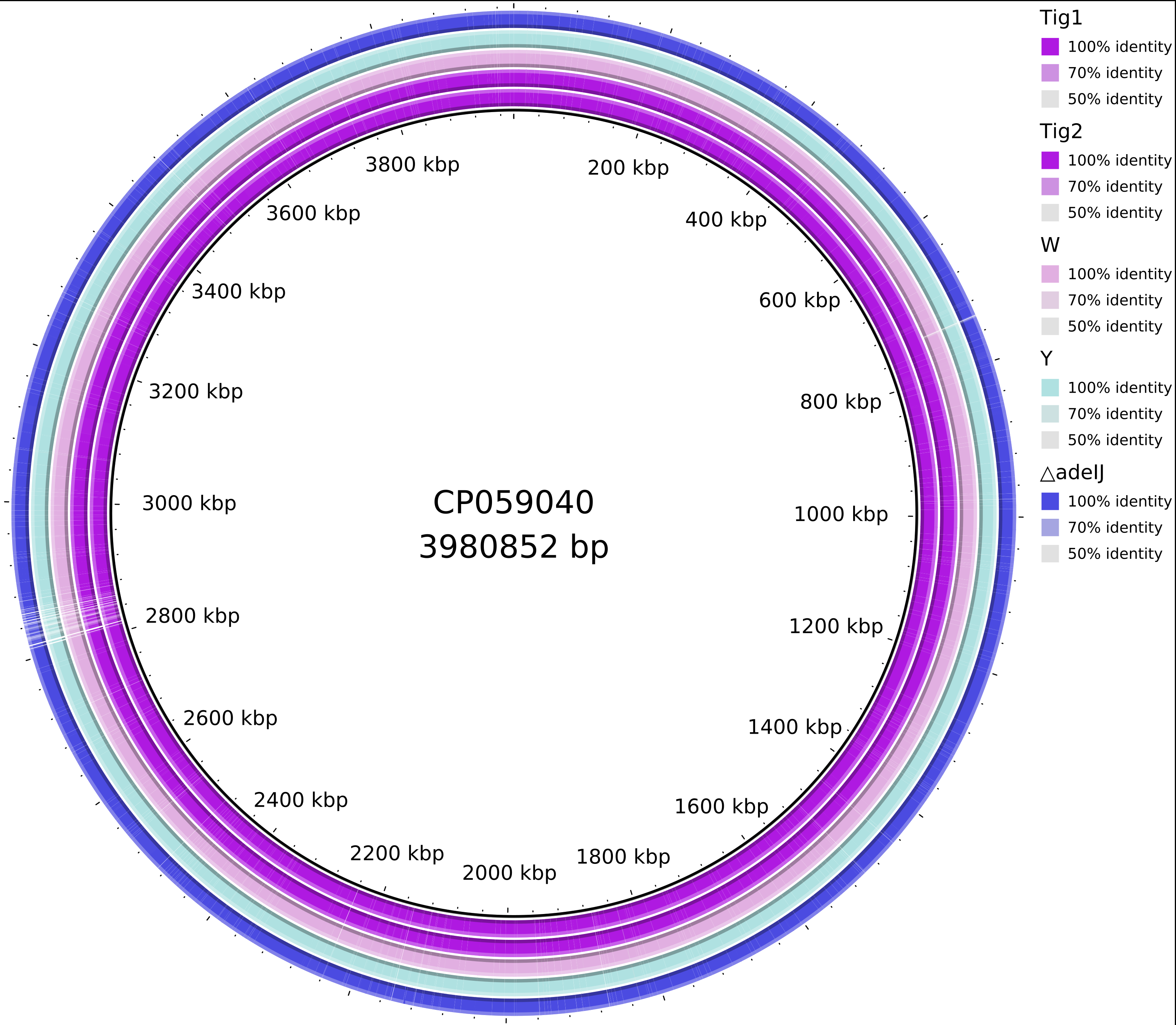
8, show difference between the genome using BRIG
This is an issue with java version. I was running Java 1.8 version and had same problem as the OP's question. Then I followed the answers and finally changing the java version worked for me.
I ran the following steps:
1) Go to link: https://www.oracle.com/java/technologies/javase-java-archive-javase6-downloads.html
2) mkdir Java1.6
3) Download "jdk-6u45-linux-x64.bin" and save in Java1.6
3) cd Java1.6 && chmod +x jdk-6u45-linux-x64.bin && ./jdk-6u45-linux-x64.bin
4) You should see java file in Java1.6/jdk1.6.0_45/bin
5) Java1.6/jdk1.6.0_45/bin/java -version - should give "1.6.0_45"
6) (base) conda install bioconda::blast-legacy # install blastall 2.2.26
7) #set lastLocation="/home/jhuang/miniconda3/bin/" in default-BRIG.xml --> /home/jhuang/miniconda3/bin/blastall is used for blast!
8) Open BRIG using Java1.6 as follows: ~/Tools/Java1.6/jdk1.6.0_45/bin/java -Xmx15000M -jar BRIG.jar
Note: I did not uninstall my original Java1.8 rather used the downloded java1.6 execulatable file using path. That way I retain both versions without any problem.
This gave me all the rings without any issue!! Hope this helps someone!
Users who wish to run BRIG from the command-line need to:
* Navigate to the unpacked BRIG folder in a command-line interface (terminal, console, command prompt).
* Run 'java -Xmx1500M -jar BRIG.jar'. Where -Xmx specifies the amount of memory allocated to BRIG.
Note:
* BLAST legacy comes as a compressed package, which will unzip the BLAST binaries where ever the package is. We advise users to first create a BLAST directory (in either the home or applications directory), copy the downloaded BLAST package to that directory and unzip the package.
* BRIG supports both BLAST+ & BLAST Legacy. If BRIG cannot find BLAST it will prompt users at runtime. Users can specify the location of their BLAST installation in the BRIG options menu which is: Main window > Preferences > BRIG options.
* N.B: If BOTH BLAST+ and legacy versions are in the same location, BRIG will prefer BLAST+
convert CP059040.png -crop 4000x2780+3840+0 CP059040_.png
9, try to use bactopia
# -- ERROR --> Aborted during installation! --
mamba create -y -n bactopia -c conda-forge -c bioconda bactopia
conda activate bactopia
bactopia datasets
# Paired-end
bactopia --R1 R1.fastq.gz --R2 R2.fastq.gz --sample SAMPLE_NAME \
--datasets datasets/ --outdir OUTDIR
# Single-End
bactopia --SE SAMPLE.fastq.gz --sample SAMPLE --datasets datasets/ --outdir OUTDIR
# Multiple Samples
bactopia prepare MY-FASTQS/ > fastqs.txt
bactopia --fastqs fastqs.txt --datasets datasets --outdir OUTDIR
# Single ENA/SRA Experiment
bactopia --accession SRX000000 --datasets datasets --outdir OUTDIR
# Multiple ENA/SRA Experiments
bactopia search "staphylococcus aureus" > accessions.txt
bactopia --accessions accessions.txt --dataset datasets --outdir ${OUTDIR}
10, IS elments using ISEScan
(base) isescan.py --seqfile Tig1.fna --output Tig1_isescan_out --nthread 20
isescan.py --seqfile Tig2.fna --output Tig2_isescan_out --nthread 20
isescan.py --seqfile Y.fna --output Y_isescan_out --nthread 20
isescan.py --seqfile W.fna --output W_isescan_out --nthread 20
isescan.py --seqfile _adeIJ.fna --output deltaAdeIJ_isescan_out --nthread 20
~/Tools/csv2xls-0.4/csv_to_xls.py ./Tig1_isescan_out/Tig1.fna.csv ./Tig2_isescan_out/Tig2.fna.csv ./Y_isescan_out/Y.fna.csv ./W_isescan_out/W.fna.csv ./deltaAdeIJ_isescan_out/_adeIJ.fna.csv -d',' -o ISEScan_res.xls
#extracted sequence segments from the two isolates, specifically:
# ATCC19606: 930469 to 951674 — segment1
# ATCC17978: 2,934,384 to 3,000,721 — segment2
#Then, I compared the two segments and found that positions 1-11055 of segment1 mapped to 66338-55284 of segment2, and positions 11049-21206 of segment1 mapped to 10158-23 of segment2. This means the sequence from 10159-55283 of segment2 (about 45 kb nt) is not mapped. I then extracted the 45 kb sequence (see the attached fasta file). I attempted to detect IS elements using the tool ISEScan (https://academic.oup.com/bioinformatics/article/33/21/3340/3930124). Four ISs were detected (see 45kb.fasta.xlsx; for more detailed results, see 45kb.fasta.zip).
#samtools faidx Acinetobacter_baumannii_ATCC19606.gbk_converted.fna CP059040.1:930469-951674 > ../ATCC19606_segment.fasta
vim ./gbks/A.baumannii_ATCC17978.gbk
#LOCUS CP000521 3976747 bp DNA circular BCT 31-JAN-2014
#DEFINITION Acinetobacter baumannii ATCC 17978, complete genome.
I used the following commands extracted a 45kb fasta. Then using a tools get IS elements.
samtools faidx A.baumannii_ATCC17978.gbk_converted.fna CP000521.1:2934384-3000721 > ../ATCC17978_segment.fasta
makeblastdb -in ATCC17978_segment.fasta -dbtype nucl
blastn -db ATCC17978_segment.fasta -query ATCC19606_segment.fasta -num_threads 15 -outfmt 6 -strand both -evalue 0.1 > ATCC19606_segment_on_ATCC17978_segment.blastn
samtools faidx ATCC17978_segment.fasta CP000521.1_2934384_3000721:10159-55283 > 45kb.fasta
please update the following tables in which all positons referred to the 45kb sequence to the complete genome in ATCC17978.
#seqID: sequence identifier
#family: family name of IS element
#cluster: Tpase cluster
#isBegin and isEnd: genome coordinates of the predicted IS element
#isLen: length of the predicted IS element
#ncopy4is: number of predicted IS copies including full-length and partial IS copies
#start1, end1, start2, end2: genome coordinates of the IRs
#score: score of the IRs
#irId: number of identical matches in pairwise alignment of left and righ hand invered repeats
#irLen, length of inverted repeats
#nGaps: number of gaps in IRs
#orfBegin, orfEnd: genome coordinates of the predicted Tpase ORF
#strand: strand where the Tpase is
#orfLen: length of predicted Tpase ORF
#E-value: the best E-value among all IS copies for the same IS element, the smaller the better
#E-value4copy: the E-value of the reported IS copy, the smaller the better
#type: type of IS element copy, 'c' for complete IS element and 'p' for partial IS element
#ov: ov number returned by hmmer search
#tir: terminal inverted repeat sequences
seqID family cluster isBegin isEnd isLen ncopy4is start1 end1 start2 end2 score irId irLen nGaps orfBegin orfEnd strand orfLen E-value E-value4copy type ov tir
CP000521.1_2934384_3000721:10159-55283 IS5 IS5_222 5818 8737 2920 1 5818 5842 8713 8737 18 17 25 0 5931 8822 + 2892 3.3E-74 3.3E-74 c 1 TGATTAAACTTTGCGGATTAAATTG:TGATTAAATCTAATGTGTTGAATTG
CP000521.1_2934384_3000721:10159-55283 IS3 IS3_176 8745 9849 1105 1 8745 8761 9833 9849 26 15 17 0 8916 9775 - 860 9E-38 9E-38 p 1 ATTGATGATAGCCAAAA:ATTGATCCTAGCCAAAA
CP000521.1_2934384_3000721:10159-55283 IS5 IS5_226 9983 10411 429 1 9983 9996 10398 10411 20 12 14 0 9850 10364 + 515 7.2E-28 7.2E-28 p 1 TATCATTCATTATA:TATCATTCAGCATA
CP000521.1_2934384_3000721:10159-55283 IS5 IS5_302 23918 24796 879 1 23918 23953 24762 24796 54 33 36 1 23947 24699 - 753 3E-82 3E-82 c 1 AAAATCAAAATAATGCTTAGGGCGTGTCCTCATTTG:AAAATCAAAATGATGC-TAGGGCGTGTCTTCATTTG
11, transposons
Insertion sequences (IS) and transposon elements are both types of mobile genetic elements, but they are distinct from each other.
Insertion Sequences (IS):
Insertion sequences are the simplest type of transposable element.
They typically consist of a short DNA sequence that encodes only the proteins necessary for their own transposition.
An IS element usually includes a transposase gene flanked by inverted repeats, which are short, repeated sequences that are necessary for the insertion process.
IS elements do not carry additional genes other than those required for their own mobility, such as antibiotic resistance genes.
Transposons:
Transposons, or transposable elements, are more complex and can be divided into two main categories: insertion sequences and composite transposons.
Composite transposons consist of two IS elements flanking a central region that often contains additional genes, such as those conferring antibiotic resistance or other functional traits.
Transposons generally include the genes required for their movement (transposase) and can carry additional genes unrelated to transposition.
Key Differences:
Insertion Sequences are the simplest type of transposon and consist of only the basic components necessary for transposition.
Transposons can include IS elements as part of their structure, but they also often contain additional genes.
So, to answer your question: Insertion Sequences do not necessarily "contain" transposon elements, but they are a type of transposon themselves. More complex transposons might include IS elements as part of their structure, along with additional genes.
12, transposon detection tools
There are several tools designed to identify and analyze transposons in genomic sequences, similar to how ISEScan is used for identifying insertion sequences (IS). Here are some popular tools for transposon detection:
* RepeatMasker (https://github.com/rmhubley/RepeatMasker/blob/master/RepeatMasker):
- Description: RepeatMasker is a widely used tool for identifying and masking repetitive elements in genomic sequences. It can detect various types of transposable elements, including both Class I (retrotransposons) and Class II (DNA transposons) elements.
- Website: RepeatMasker
* Transposome:
- Description: Transposome is a tool designed to detect and annotate transposable elements in genomic sequences. It integrates with various databases and can provide detailed information about the transposable elements found.
- Website: Transposome
* TEscan:
- Description: TEScan is a tool specifically designed for the identification and annotation of transposable elements in genomic sequences. It uses a database of transposable elements to scan the genome.
- Website: TEscan
* RepeatModeler:
- Description: RepeatModeler is used for de novo repeat identification and annotation. It helps in building custom repeat libraries and can identify transposable elements not present in pre-existing databases.
- Website: RepeatModeler
* DIGGER:
- Description: DIGGER is a tool for the detection and characterization of transposable elements in eukaryotic genomes. It uses a combination of sequence similarity searches and structural analysis.
- Website: DIGGER
* TEannotator:
- Description: TEannotator provides comprehensive annotation of transposable elements by combining different prediction methods and databases.
- Website: TEannotator
* MITE-Hunter:
- Description: MITE-Hunter is specialized for identifying miniature inverted-repeat transposable elements (MITEs), which are a subset of transposable elements.
- Website: MITE-Hunter
Each of these tools has its own strengths and may be suited to different types of analyses or types of genomes. The choice of tool might depend on the specific needs of your analysis, such as the type of transposable elements you are interested in, the complexity of your genome, and the level of detail required.
conda create --name transposons #python=3.9
conda activate transposons
# TODO: send the results and fasta files of the 5 strains so that he can do some own analysis if necessary.
1. https://github.com/Dfam-consortium/TETools
Dfam TE Tools includes RepeatMasker, RepeatModeler, and coseg. This container is an easy way to get a minimal yet fully functional installation of RepeatMasker and RepeatModeler and is additionally useful for testing or reproducibility purposes.
1. RepeatMasker
# Install RepeatMasker
mamba install -c bioconda repeatmasker
# Run RepeatMasker
RepeatMasker -species "species_name" input_file.fasta
# Replace "species_name" with the target species and input_file.fasta with your genomic sequence file.
RepeatMasker -species "baumannii" Tig1.fna --output Tig1_repeatmasker_out
#https://www.biostars.org/p/215531/
#https://www.repeatmasker.org/RepeatMasker/
#https://github.com/Dfam-consortium/FamDB
#https://www.dfam.org/releases/Dfam_3.8/families/
#https://www.dfam.org/releases/Dfam_3.8/families/FamDB/README.txt
Acinetobacter baumannii
To find the transposons in Acinetobacter baumannii using FamDB, you should download the partition that includes bacterial data. Based on the partition information provided:
Partition 0 [dfam38_full.0.h5]: This partition includes Bacteria.
Steps to Download and Use Partition 0
Download Partition 0: wget https://dfam.org/releases/Dfam_3.8/famdb/dfam38_full.0.h5
Move the downloaded file to the correct directory: mv dfam38_full.0.h5 /home/jhuang/Tools/RepeatMasker/Libraries/famdb
Verify the file is in the correct location: ls /home/jhuang/Tools/RepeatMasker/Libraries/famdb
Run the famdb.py script using the downloaded partition:
./famdb.py --path /home/jhuang/Tools/RepeatMasker/Libraries/famdb/dfam38_full.0.h5
./famdb.py -i /home/jhuang/Tools/RepeatMasker/Libraries/famdb/ names baumanni
./famdb.py -i /home/jhuang/Tools/RepeatMasker/Libraries/famdb/ info
./famdb.py -i /home/jhuang/Tools/RepeatMasker/Libraries/famdb/ info
./famdb.py -i /home/jhuang/Tools/RepeatMasker/Libraries/famdb/ lineage -ad --format totals 470
./famdb.py -i /home/jhuang/Tools/RepeatMasker/Libraries/famdb/ lineage -ad --format totals "Acinetobacter baumannii"
1342 entries in ancestors; 52 lineage-specific entries; found in partitions: 0;
wget https://dfam.org/releases/Dfam_3.8/famdb/dfam38_full.0.h5 -P /home/jhuang/Tools/RepeatMasker/Libraries/famdb/
wget https://dfam.org/releases/Dfam_3.8/famdb/dfam38_full.0.h5 -P /home/jhuang/Tools/RepeatMasker/Libraries/famdb/
./famdb.py -i /home/jhuang/Tools/RepeatMasker/Libraries/famdb/ names "bacteria"
Thus, Acinetobacter baumannii does not belong to any of the partitions listed (which include Bacteria, Bacilli, Bacillaeota, Firmicutes, and Terrabacteria group). Instead, it belongs to the partition containing the Proteobacteria phylum.
./famdb.py -i /home/jhuang/Tools/RepeatMasker/Libraries/famdb/ names "Acinetobacter"
Verify the Installation and Usage
Ensure the path provided to famdb.py points to the exact location of dfam38_full.0.h5. Adjust the script command if needed, and confirm the file permissions to avoid access issues.
By following these steps, you should be able to use the FamDB database partition that contains bacterial transposons data to analyze Acinetobacter baumannii.
#https://bioinformatics.stackexchange.com/questions/2373/is-it-wise-to-use-repeatmasker-on-prokaryotes
# align genome against itself
nucmer --maxmatch --nosimplify genome.fasta genome.fasta
# select repeats and convert the corrdinates to bed format
show-coords -r -T -H out.delta | awk '{if ($1 != $3 && $2 != $4) print $0}' | awk '{print $8"\t"$1"\t"$2}' > repeats.bed
# mask those bases with bedtools
bedtools maskfasta -fi genome.fasta -bed repeats.bed -fo masked.fasta
https://bioinformatics.stackexchange.com/questions/2373/is-it-wise-to-use-repeatmasker-on-prokaryotes
https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-015-0654-5
Detection and Characterization of Transposons in Bacteria
https://pubmed.ncbi.nlm.nih.gov/31584155/
2. TnCentral: a Prokaryotic Transposable Element Database and Web Portal for Transposon Analysis
https://journals.asm.org/doi/10.1128/mbio.02060-21
https://tncentral.ncc.unesp.br/tnfinder/
usage: Tn3+TA_finder.py [-h] [-v] -f Sequences.fasta [Sequences.fasta ...] [-o Directory] [-g]
[-t cores] [-p percentage] [-c percentage] [-d base pairs] [-m]
[-e base pairs]
~/Tools/tncomp_finder/TnComp_finder.py
usage: TnComp_finder.py [-h] [-v] -f sequences.fasta [sequences.fasta ...] [-o directory]
[-p threads] [-g] [-i %] [-c %] [-d bp] [-e bp] [-k] [-s | -t]
TnComp_finder.py: error: the following arguments are required: -f/--files
#Tn3 transposon/toxin finder
~/Tools/tn3-ta_finder/Tn3+TA_finder.py -f Tig1.fna -t 100 -o Tig1_Tn3_out
~/Tools/tn3-ta_finder/Tn3+TA_finder.py -f Tig2.fna -t 100 -o Tig2_Tn3_out
~/Tools/tn3-ta_finder/Tn3+TA_finder.py -f Y.fna -t 100 -o Y_Tn3_out
~/Tools/tn3-ta_finder/Tn3+TA_finder.py -f W.fna -t 100 -o W_Tn3_out
~/Tools/tn3-ta_finder/Tn3+TA_finder.py -f _adeIJ.fna -t 30 -o deltaAdeIJ_Tn3_out
#composite transposon finder
~/Tools/tncomp_finder/TnComp_finder.py -f Tig1.fna -p 100 -o Tig1_TnComp_out
~/Tools/tncomp_finder/TnComp_finder.py -f Tig2.fna -p 100 -o Tig2_TnComp_out
~/Tools/tncomp_finder/TnComp_finder.py -f Y.fna -p 100 -o Y_TnComp_out
~/Tools/tncomp_finder/TnComp_finder.py -f W.fna -p 100 -o W_TnComp_out
~/Tools/tncomp_finder/TnComp_finder.py -f _adeIJ.fna -p 100 -o deltaAdeIJ_TnComp_out
find . -type f -empty -delete
#~/Tools/csv2xls-0.4/csv_to_xls.py Tig1_contig13.tblastn Tig1_contig19.tblastn Tig1_contig2.tblastn Tig1_contig6.tblastn Tig1_contig17.tblastn _Tig1_contig1.tblastn Tig1_contig3.tblastn Tig1_contig7.tblastn -d$'\t' -o Tig1_Tn3_finder_res.xls
#~/Tools/csv2xls-0.4/csv_to_xls.py Tig2_contig11.tblastn Tig2_contig16.tblastn _Tig2_contig1.tblastn Tig2_contig6.tblastn Tig2_contig12.tblastn Tig2_contig17.tblastn Tig2_contig2.tblastn Tig2_contig7.tblastn Tig2_contig13.tblastn Tig2_contig18.tblastn Tig2_contig3.tblastn -d$'\t' -o Tig2_Tn3_finder_res.xls
#~/Tools/csv2xls-0.4/csv_to_xls.py Y_contig12.tblastn Y_contig14.tblastn Y_contig18.tblastn _Y_contig1.tblastn Y_contig3.tblastn Y_contig8.tblastn Y_contig13.tblastn Y_contig17.tblastn Y_contig19.tblastn Y_contig2.tblastn Y_contig7.tblastn -d$'\t' -o Y_Tn3_finder_res.xls
#~/Tools/csv2xls-0.4/csv_to_xls.py W_contig13.tblastn _W_contig1.tblastn W_contig3.tblastn W_contig7.tblastn W_contig18.tblastn W_contig20.tblastn W_contig6.tblastn -d$'\t' -o W_Tn3_finder_res.xls
#~/Tools/csv2xls-0.4/csv_to_xls.py _adeIJ_contig13.tblastn _adeIJ_contig19.tblastn _adeIJ_contig3.tblastn _adeIJ_contig7.tblastn _adeIJ_contig17.tblastn __adeIJ_contig1.tblastn _adeIJ_contig6.tblastn -d$'\t' -o deltaAdeIJ_Tn3_finder_res.xls
#~/Tools/csv2xls-0.4/csv_to_xls.py Tig1_TnComp_out/blastn/Tig1.blastn Tig2_TnComp_out/blastn/Tig2.blastn Y_TnComp_out/blastn/Y.blastn W_TnComp_out/blastn/W.blastn deltaAdeIJ_TnComp_out/blastn/_adeIJ.blastn -d$'\t' -o TnComp_finder_res.xls
#shorten the sheet names of the generated xls-file.
3. Transposome
# Clone the Transposome repository
git clone https://github.com/sestaton/Transposome.git
cd Transposome
# Install
sudo apt-get install -y build-essential lib32z1 git ncbi-blast+ curl
#The command curl -L cpanmin.us | perl - --installdeps . is used to install the Perl module dependencies specified in the current directory (typically a Makefile.PL or Build.PL file). By default, cpanm (App::cpanminus) installs Perl modules in the system Perl library directories. The exact location depends on the system and how Perl is configured. Common locations include: /usr/local/lib/perl5/, /usr/lib/perl5/, or ~/perl5/lib/perl5/
#/home/jhuang/miniconda3/envs/transposons/bin/perl
#The 70 perl libraries are installed under ~/miniconda3/envs/transposons/lib/perl5/site_perl
curl -L cpanmin.us | perl - --installdeps .
perl Makefile.PL
make
make test
make install
# Run Transposome
transposome run --config config_file.yml
# Create a configuration file config_file.yml with appropriate parameters and input files.
4. TEscan
# Install from Bioconda
conda install -c bioconda teannot
# Run TEannot with TEScan
TEannot -i Tig1.fna -o Tig1_TEScan_out
# Replace input_file.fasta with your genomic sequence file and specify an output_directory.
5. RepeatModeler
# Install RepeatModeler
conda install -c bioconda repeatmodeler
# Build a database for the input genome
BuildDatabase -name genomeDB input_file.fasta
# Run RepeatModeler
RepeatModeler -database genomeDB -pa 4
# Replace input_file.fasta with your genomic sequence file and 4 with the number of CPUs to use.
6. DIGGER
# Clone the DIGGER repository
git clone https://github.com/rosenb/digger.git
cd digger
# Install dependencies using conda
conda create -n digger -c bioconda biopython blast
conda activate digger
# Run DIGGER
python digger.py -i input_file.fasta -d database_file -o output_directory
# Replace input_file.fasta with your genomic sequence file, database_file with a suitable transposon database, and specify an output_directory.
7. TEannotator
# Clone the TEannotator repository
git clone https://github.com/teannotator/TEannotator.git
cd TEannotator
# Install dependencies using conda
conda create -n teannotator -c bioconda biopython blast
conda activate teannotator
# Run TEannotator
python TEannotator.py -i input_file.fasta -d database_file -o output_directory
# Replace input_file.fasta with your genomic sequence file, database_file with a suitable transposon database, and specify an output_directory.
8. MITE-Hunter
# Download MITE-Hunter
wget http://target.iplantcollaborative.org/ufloader/MITE_Hunter.tar.gz
tar -xzf MITE_Hunter.tar.gz
cd MITE_Hunter
# Install dependencies using conda
conda create -n mitehunter -c bioconda biopython
conda activate mitehunter
# Run MITE-Hunter
perl MITE_Hunter_manager.pl -i input_file.fasta -g genome_size -c 10
# Replace input_file.fasta with your genomic sequence file and genome_size with the estimated genome size.
13, draw the chromosome comparisons using BRIG, java version causing errors
<?xml version="1.0" encoding="UTF-8"?>
<BRIG blastOptions="" legendPosition="upper-right" queryFile="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/db/CP059040.gb_converted.fna" outputFolder="/home/jhuang/DATA/Data_Tam_variant_calling/brig_out" blastPlus="yes" outputFile="/home/jhuang/DATA/Data_Tam_variant_calling/brig_out/xxxxxx" title="CP059040" imageFormat="svg" queryFastaFile="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/db/CP059040.gb_converted.fna" cgXML="/home/jhuang/DATA/Data_Tam_variant_calling/brig_out/scratch/CP059040.gb_converted.fna.xml">
<cgview_settings arrowheadLength="medium" backboneColor="black" backboneRadius="700" backboneThickness="medium" backgroundColor="white" borderColor="black" featureSlotSpacing="medium" featureThickness="30" giveFeaturePositions="false" globalLabel="true" height="4500" isLinear="false" labelFont="SansSerif,plain,30" labelLineLength="medium" labelLineThickness="medium" labelPlacementQuality="best" labelsToKeep="1000" longTickColor="black" minimumFeatureLength="medium" moveInnerLabelsToOuter="true" origin="12" rulerFont="SansSerif,plain,35" rulerFontColor="black" rulerPadding="40" rulerUnits="bases" shortTickColor="black" shortTickThickness="medium" showBorder="true" showShading="true" showWarning="false" tickDensity="0.2333" tickThickness="medium" titleFont="SansSerif,plain,55" titleFontColor="black" useColoredLabelBackgrounds="false" useInnerLabels="true" warningFont="Default,plain,35" warningFontColor="black" width="4500" zeroTickColor="black" />
<brig_settings Ring1="172,14,225" Ring2="222,149,220" Ring3="161,221,231" Ring4="49,34,221" Ring5="116,152,226" Ring6="224,206,38" Ring7="40,191,140" Ring8="158,223,139" Ring9="226,38,122" Ring10="211,41,77" defaultUpper="70" defaultLower="50" defaultMinimum="50" genbankFiles="gbk,gb,genbank" fastaFiles="fna,faa,fas,fasta,fa" emblFiles="embl" blastLocation="" divider="3" multiplier="3" memory="1500" defaultSpacer="0" />
<special value="GC Content" />
<special value="GC Skew" />
<refDir location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/Tig1">
<refFile location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/Tig1/contigs.fa" />
</refDir>
<refDir location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/Tig2">
<refFile location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/Tig2/contigs.fa" />
</refDir>
<refDir location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/Y">
<refFile location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/Y/contigs.fa" />
</refDir>
<refDir location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/W">
<refFile location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/W/contigs.fa" />
</refDir>
<refDir location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/_adeIJ">
<refFile location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/_adeIJ/contigs.fa" />
</refDir>
<ring position="0" colour="172,14,225" name="Tig1" upperInt="70" lowerInt="50" legend="yes" size="30" labels="yes" blastType="blastn">
<sequence location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/Tig1/contigs.fa" blastResults="/home/jhuang/DATA/Data_Tam_variant_calling/brig_out/scratch/contigs.faVsCP059040.gb_converted.fna.tab" />
</ring>
<ring colour="172,14,225" name="Tig2" position="1" upperInt="70" lowerInt="50" legend="yes" size="30" labels="yes" blastType="blastn">
<sequence location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/Tig2/contigs.fa" blastResults="/home/jhuang/DATA/Data_Tam_variant_calling/brig_out/scratch/contigs.faVsCP059040.gb_converted.fna.tab" />
</ring>
<ring colour="222,149,220" name="Y" position="2" upperInt="70" lowerInt="50" legend="yes" size="30" labels="yes" blastType="blastn">
<sequence location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/Y/contigs.fa" blastResults="/home/jhuang/DATA/Data_Tam_variant_calling/brig_out/scratch/contigs.faVsCP059040.gb_converted.fna.tab" />
</ring>
<ring colour="161,221,231" name="W" position="3" upperInt="70" lowerInt="50" legend="yes" size="30" labels="no" blastType="blastn">
<sequence location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/W/contigs.fa" blastResults="/home/jhuang/DATA/Data_Tam_variant_calling/brig_out/scratch/contigs.faVsCP059040.gb_converted.fna.tab" />
</ring>
<ring colour="49,34,221" name="△adeIJ" position="4" upperInt="70" lowerInt="50" legend="yes" size="30" labels="yes" blastType="blastn">
<sequence location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/_adeIJ/contigs.fa" blastResults="/home/jhuang/DATA/Data_Tam_variant_calling/brig_out/scratch/contigs.faVsCP059040.gb_converted.fna.tab" />
</ring>
<ring colour="225,0,0" name="GC Content" position="5" upperInt="70" lowerInt="50" legend="yes" size="30" labels="no" blastType="blastn">
<sequence location="GC Content" />
</ring>
<ring colour="225,0,0" name="GC Skew" position="6" upperInt="70" lowerInt="50" legend="yes" size="30" labels="yes" blastType="blastn">
<sequence location="GC Skew" />
</ring>
</BRIG>
14, (optional) MANUALLY REMOVE the column f6 in filtered_merged_variants_CP133676.csv, and rename CHROM to HDRNA_01_K01 in the header, summarize chr and plasmids SNPs of a sample together to a single list, save as an Excel-file.
#TOMORROW_TODO_FROM_HERE, generate local genetic environments of adeIJ (Figure_1); make the variant_calling.xls; search all SNP, InDel,transposons, IS elements using http://xgenes.com/article/article-content/316/insertion-sequence-is-element-detection/, however, we have to align the found positions of Treffer; generate a BRIG using Tig1 as the reference, Tig2, delta_adeIJ, Y, W to look if 99% are idential (Figure_2).
# I am not sure what do you mean about transposons. Actually I now processed a project of the Transposons (search for the manuscript of Patricia). What do you mean? They have some specifial expected transposons so that I can check the positions of pre-defined transponsons by comparing the genomes (see the slide of Patrick)?
#TODO: Using the method using for Patricia, I can compare the five genomes to REF to look for the positions newly inserted.
15, code of summarize_snippy_res.py
import pandas as pd
import glob
import argparse
import os
#python3 summarize_snps_indels.py snippy_HDRNA_01/snippy
#The following record for 2365295 is wrong, since I am sure in the HDRNA_01_K010, it should be a 'G', since in HDRNA_01_K010.csv
#CP133676,2365295,snp,A,G,G:178 A:0
#
#The current output is as follows:
#CP133676,2365295,A,snp,A,A,A,A,A,A,A,A,A,A,None,,,,,,None,None
#CP133676,2365295,A,snp,A,A,A,A,A,A,A,A,A,A,nan,,,,,,nan,nan
#grep -v "None,,,,,,None,None" summary_snps_indels.csv > summary_snps_indels_.csv
#BUG: CP133676,2365295,A,snp,A,A,A,A,A,A,A,A,A,A,nan,,,,,,nan,nan
import pandas as pd
import glob
import argparse
import os
def main(base_directory):
# List of isolate identifiers
#isolates = [f"HDRNA_01_K0{i}" for i in range(1, 11)]
isolates = ["Tig1", "Tig2", "Y", "W", "_adeIJ"]
expected_columns = ["CHROM", "POS", "REF", "ALT", "TYPE", "EFFECT", "LOCUS_TAG", "GENE", "PRODUCT"]
# Find all CSV files in the directory and its subdirectories
csv_files = glob.glob(os.path.join(base_directory, '**', '*.csv'), recursive=True)
# Create an empty DataFrame to store the summary
summary_df = pd.DataFrame()
# Iterate over each CSV file
for file_path in csv_files:
# Extract isolate identifier from the file name
isolate = os.path.basename(file_path).replace('.csv', '')
df = pd.read_csv(file_path)
# Ensure all expected columns are present, adding missing ones as empty columns
for col in expected_columns:
if col not in df.columns:
df[col] = None
# Extract relevant columns
df = df[expected_columns]
# Ensure consistent data types
df = df.astype({"CHROM": str, "POS": int, "REF": str, "ALT": str, "TYPE": str, "EFFECT": str, "LOCUS_TAG": str, "GENE": str, "PRODUCT": str})
# Add the isolate column with the ALT allele
df[isolate] = df["ALT"]
# Drop columns that are not needed for the summary
df = df.drop(["ALT"], axis=1)
if summary_df.empty:
summary_df = df
else:
summary_df = pd.merge(summary_df, df, on=["CHROM", "POS", "REF", "TYPE", "EFFECT", "LOCUS_TAG", "GENE", "PRODUCT"], how="outer")
# Fill missing values with the REF allele for each isolate column
for isolate in isolates:
if isolate in summary_df.columns:
summary_df[isolate] = summary_df[isolate].fillna(summary_df["REF"])
else:
summary_df[isolate] = summary_df["REF"]
# Rename columns to match the required format
summary_df = summary_df.rename(columns={
"CHROM": "CHROM",
"POS": "POS",
"REF": "REF",
"TYPE": "TYPE",
"EFFECT": "Effect",
"LOCUS_TAG": "Gene_name",
"GENE": "Biotype",
"PRODUCT": "Product"
})
# Replace any remaining None or NaN values in the non-isolate columns with empty strings
summary_df = summary_df.fillna("")
# Add empty columns for Impact, Functional_Class, Codon_change, Protein_and_nucleotide_change, Amino_Acid_Length
summary_df["Impact"] = ""
summary_df["Functional_Class"] = ""
summary_df["Codon_change"] = ""
summary_df["Protein_and_nucleotide_change"] = ""
summary_df["Amino_Acid_Length"] = ""
# Reorder columns
cols = ["CHROM", "POS", "REF", "TYPE"] + isolates + ["Effect", "Impact", "Functional_Class", "Codon_change", "Protein_and_nucleotide_change", "Amino_Acid_Length", "Gene_name", "Biotype"]
summary_df = summary_df.reindex(columns=cols)
# Remove duplicate rows
summary_df = summary_df.drop_duplicates()
# Save the summary DataFrame to a CSV file
output_file = os.path.join(base_directory, "summary_snps_indels.csv")
summary_df.to_csv(output_file, index=False)
print("Summary CSV file created successfully at:", output_file)
if __name__ == "__main__":
parser = argparse.ArgumentParser(description="Summarize SNPs and Indels from CSV files.")
parser.add_argument("directory", type=str, help="Base directory containing the CSV files in subdirectories.")
args = parser.parse_args()
main(args.directory)
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Kraken2 Installation and Usage Guide
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Should the inputs for GSVA be normalized or raw?
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- Risks of Rebooting into Rescue Mode
- 足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Structural Variant Calling for Nanopore Sequencing (edited)
Call and merge SNP and Indel results from using two variant calling methods
Scaffolding and finishing an assembly with a reference genome