Typing of S. epidermidis samples (HDMx samples)
gene_x 0 like s 808 view s
Tags: processing
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2786320/
Classification of Staphylococcal Cassette Chromosome mec (SCCmec): Guidelines for Reporting Novel SCCmec Elements
mec gene complex.
IS1272 is part of the IS1182 family of insertion sequences
- The mec gene complex is composed of mecA, its regulatory genes, and associated insertion sequences.
- The class A mec gene complex (class A mec) is the prototype complex, which contains mecA, the complete mecR1 and mecI regulatory genes upstream of mecA, and the hypervariable region (HVR) and insertion sequence IS431 downstream of mecA.
-
- The class B mec gene complex is composed of mecA, a truncated mecR1 resulting from the insertion of IS1272 upstream of mecA, and [HVR and IS431 downstream of mecA].
- The class C mec gene complex contains mecA and truncated mecR1 by the insertion of IS431 upstream of mecA and HVR and IS431 downstream of mecA.
- There are two distinct class C mec gene complexes; in the class C1 mec gene complex, the IS431 upstream of mecA has the same orientation as the IS431 downstream of mecA (next to HVR), while in the class C2 mec gene complex, the orientation of IS431 upstream of mecA is reversed.
- C1 and C2 are regarded as different mec gene complexes since they have likely evolved independently.
- The class D mec gene complex is composed of mecA and ΔmecR1 but does not carry an insertion sequence downstream of ΔmecR1 (as determined by PCR analysis).
-
Several variants within the major classes of the mec gene complex have been described, including insertions of IS431 or IS1182 upstream of mecA in the class A mec gene complex or insertion of Tn4001 upstream of mecA in the class B mec complex.
-
run with bengal3
cd ~/DATA/Data_Denise_CalCov1 cp bacto-0.1.json ../Data_Denise_CalCov2 cp cluster.json ../Data_Denise_CalCov2 cp Snakefile ../Data_Denise_CalCov2 ln -s /home/jhuang/Tools/bacto/local . ln -s /home/jhuang/Tools/bacto/db . ln -s /home/jhuang/Tools/bacto/envs . mkdir raw_data; cd raw_data ln -s ../Alignment_Imported_1/20240913_174420/Fastq/HDM7_S1_L001_R1_001.fastq.gz HDM7_R1.fastq.gz ln -s ../Alignment_Imported_1/20240913_174420/Fastq/HDM7_S1_L001_R2_001.fastq.gz HDM7_R2.fastq.gz ln -s ../Alignment_Imported_1/20240913_174420/Fastq/HDM10_S2_L001_R1_001.fastq.gz HDM10_R1.fastq.gz ln -s ../Alignment_Imported_1/20240913_174420/Fastq/HDM10_S2_L001_R2_001.fastq.gz HDM10_R2.fastq.gz ln -s ../20240812_FS10003086_50_BSB09416-2831/Alignment_Imported_1/20240813_202730/Fastq/HDM1_S1_L001_R1_001.fastq.gz HDM1_R1.fastq.gz ln -s ../20240812_FS10003086_50_BSB09416-2831/Alignment_Imported_1/20240813_202730/Fastq/HDM1_S1_L001_R2_001.fastq.gz HDM1_R2.fastq.gz ln -s ../20240913/Alignment_Imported_1/20240913_174420/Fastq/HDM7_S1_L001_R1_001.fastq.gz HDM7_R1.fastq.gz ln -s ../20240913/Alignment_Imported_1/20240913_174420/Fastq/HDM7_S1_L001_R2_001.fastq.gz HDM7_R2.fastq.gz ln -s ../20240913/Alignment_Imported_1/20240913_174420/Fastq/HDM10_S2_L001_R1_001.fastq.gz HDM10_R1.fastq.gz ln -s ../20240913/Alignment_Imported_1/20240913_174420/Fastq/HDM10_S2_L001_R2_001.fastq.gz HDM10_R2.fastq.gz ln -s ../20240919_FS10003086_61_BSB09416-2735/Alignment_Imported_1/20240920_173408/Fastq/HDM11-SF1_S1_L001_R1_001.fastq.gz HDM11-SF1_R1.fastq.gz ln -s ../20240919_FS10003086_61_BSB09416-2735/Alignment_Imported_1/20240920_173408/Fastq/HDM11-SF1_S1_L001_R2_001.fastq.gz HDM11-SF1_R2.fastq.gz ln -s ../20240919_FS10003086_61_BSB09416-2735/Alignment_Imported_1/20240920_173408/Fastq/HDM15-SF2_S2_L001_R1_001.fastq.gz HDM15-SF2_R1.fastq.gz ln -s ../20240919_FS10003086_61_BSB09416-2735/Alignment_Imported_1/20240920_173408/Fastq/HDM15-SF2_S2_L001_R2_001.fastq.gz HDM15-SF2_R2.fastq.gz # only activate the steps assembly and mlst in bacto-0.1.json. (bengal3_ac3) jhuang@WS-2290C:~/Documents$ /home/jhuang/miniconda3/envs/snakemake_4_3_1/bin/snakemake --printshellcmds # -- Results -- shovill/HDM1/contigs.fa sepidermidis 5 arcC(1) aroE(1) gtr(1) mutS(2) pyrR(2) tpiA(1) yqiL(1) HDM10.mlst.txt:shovill/HDM10/contigs.fa sepidermidis 59 arcC(2) aroE(1) gtr(1) mutS(1) pyrR(2) tpiA(1) yqiL(1) HDM7.mlst.txt:shovill/HDM7/contigs.fa sepidermidis 59 arcC(2) aroE(1) gtr(1) mutS(1) pyrR(2) tpiA(1) yqiL(1) -
run with bakta
#under env (bakta) for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do bakta --db /mnt/nvme0n1p1/bakta_db shovill/${sample}/contigs.fa --prefix ${sample} done -
mapping on assembly to calculate the coverage
#samtools depth input.bam > depth.txt #samtools depth input.bam | awk '{sum+=$3} END { print "Average coverage:",sum/NR}' #bedtools coverage -a regions.bed -b input.bam > coverage.txt #bedtools coverage -a regions.bed -b input.bam -d > coverage_per_base.txt bwa index ./shovill/HDM1/contigs.fa bwa mem ./shovill/HDM1/contigs.fa fastq/HDM1_1.fastq fastq/HDM1_2.fastq > aligned.sam samtools view -Sb aligned.sam > aligned.bam samtools sort aligned.bam -o sorted.bam samtools index sorted.bam samtools depth sorted.bam > depth.txt awk '{sum+=$3} END { print "Average coverage:",sum/NR}' depth.txt bedtools coverage -a regions.bed -b sorted.bam > coverage.txt bedtools genomecov -ibam sorted.bam -d > coverage_per_base.txt # Step 1: Calculate depth using samtools samtools depth sorted.bam > depth.txt # Step 2: Calculate average depth using awk awk '{sum+=$3; count++} END {print "Average Coverage:", sum/count}' depth.txt # Step 1: Calculate coverage with bedtools for a BED file #bedtools coverage -a regions.bed -b input.bam > coverage.txt # Step 2: Process the output with awk #awk '{ sum+=$7 } END { print "Average coverage depth:", sum/NR }' coverage.txt bwa index ./shovill/HDM7/contigs.fa bwa mem ./shovill/HDM7/contigs.fa fastq/HDM7_1.fastq fastq/HDM7_2.fastq > aligned_HDM7.sam samtools view -Sb aligned_HDM7.sam > aligned_HDM7.bam samtools sort aligned_HDM7.bam -o sorted_HDM7.bam samtools index sorted_HDM7.bam # Step 1: Calculate depth using samtools samtools depth sorted_HDM7.bam > depth_HDM7.txt # Step 2: Calculate average depth using awk awk '{sum+=$3; count++} END {print "Average Coverage:", sum/count}' depth_HDM7.txt #Average Coverage: 380.079 bwa index ./shovill/HDM10/contigs.fa bwa mem ./shovill/HDM10/contigs.fa fastq/HDM10_1.fastq fastq/HDM10_2.fastq > aligned_HDM10.sam samtools view -Sb aligned_HDM10.sam > aligned_HDM10.bam samtools sort aligned_HDM10.bam -o sorted_HDM10.bam samtools index sorted_HDM10.bam # Step 1: Calculate depth using samtools samtools depth sorted_HDM10.bam > depth_HDM10.txt # Step 2: Calculate average depth using awk awk '{sum+=$3; count++} END {print "Average Coverage:", sum/count}' depth_HDM10.txt #Average Coverage: 254.704 -
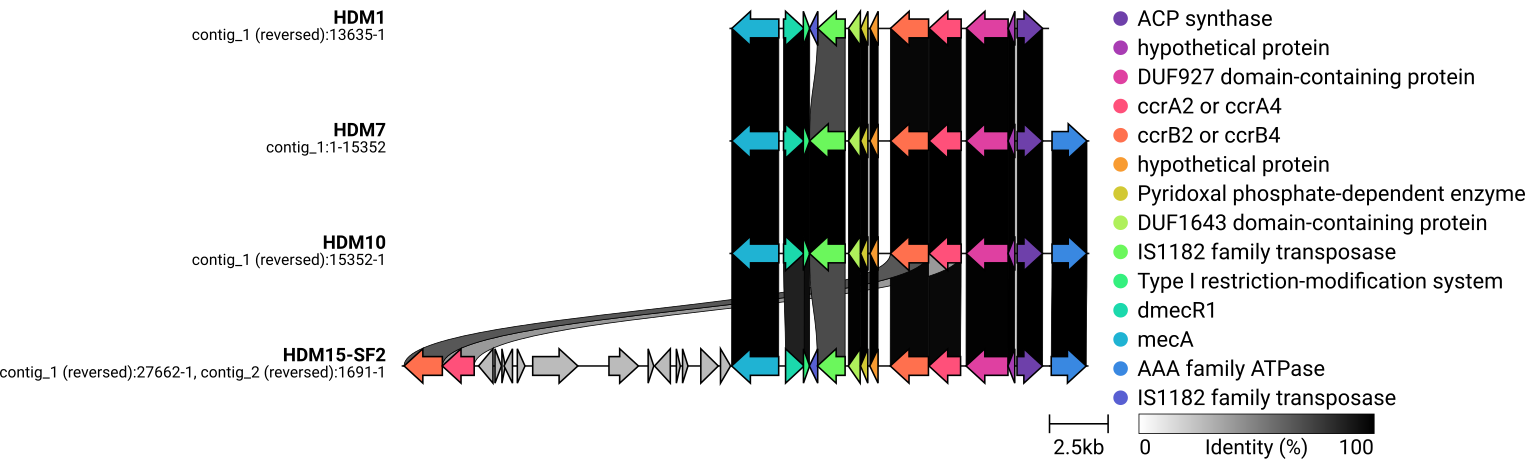
SCCmec typing and drawing with clinker
#1. -- HDM1_contigs.fa -- One SCCmec element detected. Prediction based on genes: Predicted SCCmec element: SCCmec_type_IV(2B) Prediction based on homology to whole cassette: Predicted whole cassette and %template coverage: SCCmec_type_IV(2B) 79.92% Predicted genes: Fasta header % Identity Query/HSP Length Contig Position in contig ccrA2:7:81108:AB096217 100.00 1350/1350 contig00032 3770..5119 ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00032 5120..6769 IS1272:3:AM292304 99.95 1844/1843 contig00032 8611..10454 dmecR1:1:AB033763 100.00 987/987 contig00032 10443..11429 mecA:12:AB505628 100.00 2010/2010 contig00032 11526..13535 samtools faidx shovill/HDM1_contigs.fa samtools faidx shovill/HDM1_contigs.fa contig00032:1-13635 > HDM1_sub.fna bakta --db /mnt/nvme0n1p1/bakta_db HDM1_sub.fna #2. -- HDM7_contigs.fa -- One SCCmec element detected. Prediction based on genes: Predicted SCCmec element: SCCmec_type_IVa(2B) Prediction based on homology to whole cassette: Predicted whole cassette and %template coverage: SCCmec_type_IVa(2B) 84.24% Predicted genes: Fasta header % Identity Query/HSP Length Contig Position in contig mecA:12:AB505628 100.00 2010/2010 contig00014 2800..4809 dmecR1:1:AB033763 99.90 987/987 contig00014 4906..5892 IS1272:3:AM292304 100.00 1843/1843 contig00014 5881..7723 ccrB2:3:CA05:AB063172 100.00 1629/1629 contig00014 9565..11193 ccrA2:3:CA05:AB063172 100.00 1350/1350 contig00014 11215..12564 subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00014 16461..17951 #IS1272:2:AB033763 91.06 1577/1585 contig00001 369260..370836 samtools faidx shovill/HDM7_contigs.fa samtools faidx shovill/HDM7_contigs.fa contig00014:2700-18051 > HDM7_sub.fna bakta --db /mnt/nvme0n1p1/bakta_db HDM7_sub.fna mecA dmecR1 Type I restriction enzyme HindI endonuclease subunit-like C-terminal domain-containing protein IS1272 DUF1643 domain-containing protein Pyridoxal phosphate-dependent enzyme hypothetical protein ccrB2 ccrA2 DUF927 domain-containing protein hypothetical protein ACP synthase AAA family ATPase (= subtype-IVa(2B)) #3. -- HDM10_contigs.fa -- Prediction based on genes: Predicted SCCmec element: SCCmec_type_IV(2B&5) Prediction based on homology to whole cassette: Predicted whole cassette and % template coverage: SCCmec_type_IV(2B) 84.37% Predicted genes: Fasta header % Identity Query/HSP Length Contig Position in contig subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00020 4152..5642 ccrA2:3:CA05:AB063172 100.00 1350/1350 contig00020 9539..10888 ccrB2:3:CA05:AB063172 100.00 1629/1629 contig00020 10910..12538 IS1272:3:AM292304 100.00 1843/1843 contig00020 14380..16222 dmecR1:1:AB033763 100.00 987/987 contig00020 16211..17197 mecA:12:AB505628 100.00 2010/2010 contig00020 17294..19303 #IS1272:2:AB033763 90.75 1579/1585 contig00033 2..1580 #ccrC1-allele-2:1:AB512767 90.95 1680/1680 contig00022 9836..11515 samtools faidx shovill/HDM10_contigs.fa samtools faidx shovill/HDM10_contigs.fa contig00020:4052-19403 > HDM10_sub.fna bakta --db /mnt/nvme0n1p1/bakta_db HDM10_sub.fna #4. -- HDM11-SF1_contigs.fa -- No SCCmec element was detected Prediction based on genes: Predicted SCCmec element: none Prediction based on homology to whole cassette: Predicted whole cassette and %template coverage: none #5. -- HDM15-SF2_contigs.fa -- SCCmec_type_IV(2B) SCCmec_type_VI(4B) Following gene complexes based on prediction of genes was detected : ccr class 2 ccr class 4 mec class B Predicted genes: Fasta header % Identity Query/HSP Length Contig Position in contig ccrA2:7:81108:AB096217 100.00 1350/1350 contig00004 3823..5172 ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00004 5173..6822 IS1272:3:AM292304 99.95 1844/1843 contig00004 8664..10507 dmecR1:1:AB033763 100.00 987/987 contig00004 10496..11482 mecA:12:AB505628 100.00 2010/2010 contig00004 11579..13588 subtyppe-Vc(5C2&5):10:AB505629 99.84 1935/1935 contig00004 20148..22082 ccrA4:2:BK20781:FJ670542 90.53 1362/1362 contig00004 24570..25931 ccrB4:2:BK20781:FJ670542 91.68 1635/1629 contig00004 25928..27562 subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00015 52228..53718 samtools faidx shovill/HDM7_contigs.fa samtools faidx shovill/HDM7_contigs.fa contig00014:2700-18051 > HDM7_sub.fna bakta --db /mnt/nvme0n1p1/bakta_db HDM7_sub.fna samtools faidx shovill/HDM10_contigs.fa samtools faidx shovill/HDM10_contigs.fa contig00020:4052-19403 > HDM10_sub.fna bakta --db /mnt/nvme0n1p1/bakta_db HDM10_sub.fna samtools faidx shovill/HDM15-SF2_contigs.fa samtools faidx shovill/HDM15-SF2_contigs.fa contig00004:1-27662 > HDM15-SF2_sub.fna samtools faidx shovill/HDM15-SF2_contigs.fa contig00015:52128-53818 >> HDM15-SF2_sub.fna bakta --db /mnt/nvme0n1p1/bakta_db HDM15-SF2_sub.fna #END #172.104.140.19 mkdir gbff_sub mv *_sub.gbff gbff_sub cd gbff_sub for f in *_sub.gbff; do mv "$f" "${f/_sub.gbff/.gbff}"; done #mv HDM1_sub.gbff HDM1.gbff #mv HDM7_sub.gbff HDM7.gbff #mv HDM10_sub.gbff HDM10.gbff #mv HDM15-SF2_sub.gbff HDM15-SF2.gbff rm *.json clinker *.gbff -p plot_HDRNA.html --dont_set_origin -s session_HDRNA.json -o alignments_HDRNA.csv -dl "," -dc 4 cp ./gbff_HDRNA_01/clinker.png HDRNA_01_clinker.png -
Arg typing
grep "agrD" *.gbff | sort HDM1.gbff: /gene="agrD" HDM1.gbff: /gene="agrD" HDM7.gbff: /gene="agrD" HDM7.gbff: /gene="agrD" HDM10.gbff: /gene="agrD" HDM10.gbff: /gene="agrD" HDM11-SF1.gbff: /gene="agrD" HDM11-SF1.gbff: /gene="agrD" HDM15-SF2.gbff: /gene="agrD" HDM15-SF2.gbff: /gene="agrD" MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE #* The agr typing is not defined, as I have compared the sequence with the amino acid sequences of ArgD described in the paper available at https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4187671/. It does not correspond to Type I, Type II, or Type III. (For more details, see below). -- AgrD I -- Query 1 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE 46 M + L +K F+ + IG + + C Y DEP+VPEELTKL E Sbjct 926825 MNLLGGLLLKIFSNFMAVIGNASKYNPCVMYLDEPQVPEELTKLDE 926688 -- AgrD II -- Query 1 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE 46 MNLLGGLLLKIFSNFMAVIGNASKYNPC YLDEPQVPEELTKLDE Sbjct 926825 MNLLGGLLLKIFSNFMAVIGNASKYNPCVMYLDEPQVPEELTKLDE 926688 -- AgrD III -- Query 1 MNLLGGLLLKLFSNFMAVIGNAAKYNPCASYLDEPQVPEELTKLDE 46 MNLLGGLLLK+FSNFMAVIGNA+KYNPC YLDEPQVPEELTKLDE Sbjct 926825 MNLLGGLLLKIFSNFMAVIGNASKYNPCVMYLDEPQVPEELTKLDE 926688 -
calulate the presence-absence-matrix for predefined gene list
#start codon: ATG, GTG und TTG #stop codon: 5'-UAA-3', 5'-UGA-3' und 5'-UAG-3' --> TAA, TGA, TAG ./Staphylococcus_aureus_MRSA252.fasta ./Staphylococcus_epidermidis_RP62A.fasta ./Enterococcus_faecium_isolate_E300_pathogenicity_island.fasta # -- Hause keeper: gyrB -- #samtools faidx Staphylococcus_epidermidis_RP62A.fasta "gi|57865352|ref|NC_002976.3|":2609812-2611743 > gyrB.fasta #revcomp gyrB.fasta > gyrB_revcomp.fasta gyrB_revcomp.fasta # -- Metabolic genes: fumC, gltA, icd -- #samtools faidx Staphylococcus_epidermidis_RP62A.fasta "gi|57865352|ref|NC_002976.3|":1444326-1445711 > fumC.fasta #revcomp fumC.fasta > fumC_revcomp.fasta ./fumC_revcomp.fasta ./gltA.fasta #samtools faidx Staphylococcus_epidermidis_RP62A.fasta "gi|57865352|ref|NC_002976.3|":1296195-1297463 > icd.fasta #revcomp icd.fasta > icd_revcomp.fasta icd_revcomp.fasta # -- Virulence regulartors: apsS, sigB, sarA, agrC, yycG -- #samtools faidx Staphylococcus_epidermidis_RP62A.fasta "gi|57865352|ref|NC_002976.3|":316151-317191 > apsS.fasta apsS.fasta #samtools faidx Staphylococcus_epidermidis_RP62A.fasta "gi|57865352|ref|NC_002976.3|":1722805-1723575 > sigB.fasta #revcomp sigB.fasta > sigB_revcomp.fasta sigB_revcomp.fasta #samtools faidx Staphylococcus_epidermidis_RP62A.fasta "gi|57865352|ref|NC_002976.3|":279424-279798 > sarA.fasta #revcomp sarA.fasta > sarA_revcomp.fasta sarA_revcomp.fasta ./agrC.fasta ./yycG.fasta # -- Toxins: psmβ1, hlb -- ./psm-beta.fasta #psm-beta1.fasta ./hlb_.fasta #./hlb.fasta # -- Biofilm formation: atlE, sdrG, sdrH, ebh, ebp, tagB -- ./atlE.fasta ./sdrG.fasta #samtools faidx Staphylococcus_epidermidis_RP62A.fasta "gi|57865352|ref|NC_002976.3|":1555024-1556469 > sdrH.fasta #revcomp sdrH.fasta > sdrH_revcomp.fasta sdrH_revcomp.fasta #samtools faidx Staphylococcus_epidermidis_RP62A.fasta "gi|57865352|ref|NC_002976.3|":1023531-1053980 > ebh.fasta #revcomp ebh.fasta > ebh_revcomp.fasta ebh_revcomp.fasta #https://www.ncbi.nlm.nih.gov/gene/?term=(Elastin-binding+protein)+AND+%22Staphylococcus+aureus%22%5Bporgn%3A__txid1282%5D #samtools faidx Staphylococcus_epidermidis_RP62A.fasta "gi|57865352|ref|NC_002976.3|":1094204-1095586 > ebpS.fasta #revcomp ebpS.fasta > ebpS_revcomp.fasta ebpS_revcomp.fasta ./tagB.fasta # -- Immune evasion & colonization: capC, sepA, dltA, fmtC, lipA, sceD, SE0760 -- ./capC.fasta #./capBCA_ywtC.fasta ./sepA.fasta #./ORF123_sepA_ORF5.fasta #samtools faidx Staphylococcus_epidermidis_RP62A.fasta "gi|57865352|ref|NC_002976.3|":503173-504630 > dltA.fasta dltA.fasta ./fmtC.fasta #samtools faidx Staphylococcus_epidermidis_RP62A.fasta "gi|57865352|ref|NC_002976.3|":498445-499359 > lipA.fasta lipA.fasta ./sceD.fasta #./sceDAE.fasta ./SE0760.fasta # -- Serine protease: esp, ecpA -- ./esp.fasta ./ecpA_.fasta #./ecpA.fasta # -- Phage: PhiSepi-HH1, PI-Sepi-HH2, PhiSepi-HH3 (#HH1-HP1, HH3-HP2, HH3-TreR) -- ./MT880870.fasta ./MT880871.fasta ./MT880872.fasta #Note that write a message to Holger, say "ebp gene does not exist, instead of it only ebpS gene exists!" makeblastdb -in HDM1_contigs.fa -dbtype nucl for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query gyrB_revcomp.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > gyrB_on_${sample}.blastn done ./fumC_revcomp.fasta ./gltA.fasta icd_revcomp.fasta for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query fumC_revcomp.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > fumC_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query gltA.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > gltA_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query icd_revcomp.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > icd_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query apsS.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > apsS_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query sigB_revcomp.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sigB_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query sarA_revcomp.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sarA_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query agrC.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > agrC_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query yycG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > yycG_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query psm-beta.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > psm-beta_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query psm-beta1.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > psm-beta1_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query hlb_.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > hlb_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query atlE.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > atlE_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query sdrG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sdrG_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query sdrH_revcomp.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sdrH_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query ebh_revcomp.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ebh_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query ebpS_revcomp.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ebpS_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query tagB.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > tagB_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query capC.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > capC_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query sepA.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sepA_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query dltA.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > dltA_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query fmtC.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > fmtC_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query lipA.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > lipA_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query sceD.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sceD_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query SE0760.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > SE0760_on_${sample}.blastn done # -- Serine protease: esp, ecpA -- for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query esp.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./esp_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query ecpA_.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ecpA_on_${sample}.blastn done # -- Phage: PhiSepi-HH1, PI-Sepi-HH2, PhiSepi-HH3 (#HH1-HP1, HH3-HP2, HH3-TreR) -- #34053 (3000,2510) 36164 (500) 147057 (6618, 15237+3812, 15237+3812, 15233+3814, 15230+3812) for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query MT880870.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./MT880870_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query MT880871.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./MT880871_on_${sample}.blastn done for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2; do blastn -db ../shovill/${sample}_contigs.fa -query MT880872.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880872_on_${sample}.blastn done
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Kraken2 Installation and Usage Guide
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Should the inputs for GSVA be normalized or raw?
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- Risks of Rebooting into Rescue Mode
- 足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
Human RNA-seq processing for Data_Ben_RNAseq_2025
Processing Data_Michelle_RNAseq_2025
From Genbank-ID to Phylogenetic Tree Visualization using roary, raxml and ete3