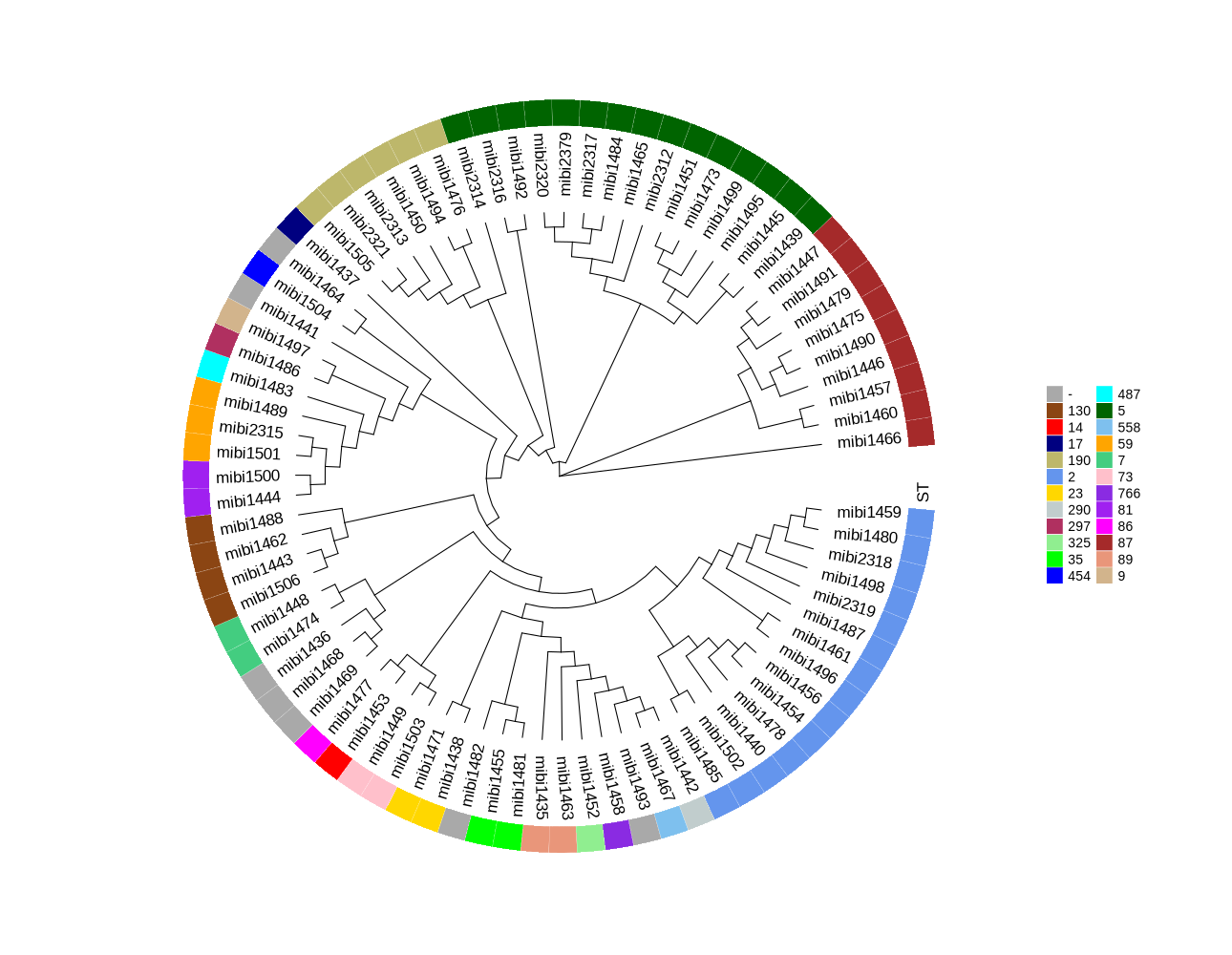
Typing of 81 S. epidermidis samples (Luise)
gene_x 0 like s 999 view s
Tags: processing, pipeline
-
prepare the bacto environment
cd ~/DATA/Data_Luise_Sepi_STKN cp /home/jhuang/Tools/bacto/bacto-0.1.json . cp /home/jhuang/Tools/bacto/cluster.json . cp /home/jhuang/Tools/bacto/Snakefile . ln -s /home/jhuang/Tools/bacto/local . ln -s /home/jhuang/Tools/bacto/db . ln -s /home/jhuang/Tools/bacto/envs . -
prepare raw_data
mkdir raw_data; cd raw_data ln -s ../raw_data_batch2/mibi2312/Luise_1_S33_R1_001.fastq.gz mibi2312_R1.fastq.gz ln -s ../raw_data_batch2/mibi2312/Luise_1_S33_R2_001.fastq.gz mibi2312_R2.fastq.gz ln -s ../raw_data_batch2/mibi2379/Luise_2_S3_R1_001.fastq.gz mibi2379_R1.fastq.gz ln -s ../raw_data_batch2/mibi2379/Luise_2_S3_R2_001.fastq.gz mibi2379_R2.fastq.gz ln -s ../raw_data_batch2/mibi2313/Luise_3_S34_R1_001.fastq.gz mibi2313_R1.fastq.gz ln -s ../raw_data_batch2/mibi2313/Luise_3_S34_R2_001.fastq.gz mibi2313_R2.fastq.gz ln -s ../raw_data_batch2/mibi2380/Luise_4_S4_R1_001.fastq.gz mibi2380_R1.fastq.gz ln -s ../raw_data_batch2/mibi2380/Luise_4_S4_R2_001.fastq.gz mibi2380_R2.fastq.gz ln -s ../raw_data_batch2/mibi2314/Luise_5_S35_R1_001.fastq.gz mibi2314_R1.fastq.gz ln -s ../raw_data_batch2/mibi2314/Luise_5_S35_R2_001.fastq.gz mibi2314_R2.fastq.gz ln -s ../raw_data_batch2/mibi2315/Luise_6_S36_R1_001.fastq.gz mibi2315_R1.fastq.gz ln -s ../raw_data_batch2/mibi2315/Luise_6_S36_R2_001.fastq.gz mibi2315_R2.fastq.gz ln -s ../raw_data_batch2/mibi2381/Luise_7_S5_R1_001.fastq.gz mibi2381_R1.fastq.gz ln -s ../raw_data_batch2/mibi2381/Luise_7_S5_R2_001.fastq.gz mibi2381_R2.fastq.gz ln -s ../raw_data_batch2/mibi2316/Luise_8_S37_R1_001.fastq.gz mibi2316_R1.fastq.gz ln -s ../raw_data_batch2/mibi2316/Luise_8_S37_R2_001.fastq.gz mibi2316_R2.fastq.gz ln -s ../raw_data_batch2/mibi2317/Luise_9_S38_R1_001.fastq.gz mibi2317_R1.fastq.gz ln -s ../raw_data_batch2/mibi2317/Luise_9_S38_R2_001.fastq.gz mibi2317_R2.fastq.gz ln -s ../raw_data_batch2/mibi2318/Luise_10_S39_R1_001.fastq.gz mibi2318_R1.fastq.gz ln -s ../raw_data_batch2/mibi2318/Luise_10_S39_R2_001.fastq.gz mibi2318_R2.fastq.gz ln -s ../raw_data_batch2/mibi2319/Luise_11_S40_R1_001.fastq.gz mibi2319_R1.fastq.gz ln -s ../raw_data_batch2/mibi2319/Luise_11_S40_R2_001.fastq.gz mibi2319_R2.fastq.gz ln -s ../raw_data_batch2/mibi2320/Luise_12_S41_R1_001.fastq.gz mibi2320_R1.fastq.gz ln -s ../raw_data_batch2/mibi2320/Luise_12_S41_R2_001.fastq.gz mibi2320_R2.fastq.gz ln -s ../raw_data_batch2/mibi2321/Luise_13_S42_R1_001.fastq.gz mibi2321_R1.fastq.gz ln -s ../raw_data_batch2/mibi2321/Luise_13_S42_R2_001.fastq.gz mibi2321_R2.fastq.gz # raw_data_batch1: mibi1435 - mibi1506 ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1435/S_epi_K_38_S1_R1_001.fastq.gz mibi1435_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1435/S_epi_K_38_S1_R2_001.fastq.gz mibi1435_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1436/S_epi_K_68_S2_R1_001.fastq.gz mibi1436_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1436/S_epi_K_68_S2_R2_001.fastq.gz mibi1436_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1437/S_epi_K_70_S3_R1_001.fastq.gz mibi1437_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1437/S_epi_K_70_S3_R2_001.fastq.gz mibi1437_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1438/S_epi_K_71_S4_R1_001.fastq.gz mibi1438_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1438/S_epi_K_71_S4_R2_001.fastq.gz mibi1438_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1439/S_epi_K_48_S5_R1_001.fastq.gz mibi1439_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1439/S_epi_K_48_S5_R2_001.fastq.gz mibi1439_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1440/S_epi_K_51_S6_R1_001.fastq.gz mibi1440_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1440/S_epi_K_51_S6_R2_001.fastq.gz mibi1440_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1441/S_epi_K_53_S7_R1_001.fastq.gz mibi1441_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1441/S_epi_K_53_S7_R2_001.fastq.gz mibi1441_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1442/S_epi_K_54_S8_R1_001.fastq.gz mibi1442_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1442/S_epi_K_54_S8_R2_001.fastq.gz mibi1442_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1443/S_epi_K_56_S9_R1_001.fastq.gz mibi1443_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1443/S_epi_K_56_S9_R2_001.fastq.gz mibi1443_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1444/S_epi_K_63_S10_R1_001.fastq.gz mibi1444_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1444/S_epi_K_63_S10_R2_001.fastq.gz mibi1444_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1445/S_epi_K_64_S11_R1_001.fastq.gz mibi1445_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1445/S_epi_K_64_S11_R2_001.fastq.gz mibi1445_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1446/S_epi_K_65_S12_R1_001.fastq.gz mibi1446_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1446/S_epi_K_65_S12_R2_001.fastq.gz mibi1446_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1447/S_epi_K_75_S13_R1_001.fastq.gz mibi1447_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1447/S_epi_K_75_S13_R2_001.fastq.gz mibi1447_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1448/S_epi_K_11_S14_R1_001.fastq.gz mibi1448_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1448/S_epi_K_11_S14_R2_001.fastq.gz mibi1448_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1449/S_epi_K_12_S15_R1_001.fastq.gz mibi1449_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1449/S_epi_K_12_S15_R2_001.fastq.gz mibi1449_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1450/S_epi_K_13_S16_R1_001.fastq.gz mibi1450_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1450/S_epi_K_13_S16_R2_001.fastq.gz mibi1450_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1451/S_epi_K_77_S17_R1_001.fastq.gz mibi1451_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1451/S_epi_K_77_S17_R2_001.fastq.gz mibi1451_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1452/S_epi_K_79_S18_R1_001.fastq.gz mibi1452_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1452/S_epi_K_79_S18_R2_001.fastq.gz mibi1452_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1453/S_epi_K_80_S19_R1_001.fastq.gz mibi1453_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1453/S_epi_K_80_S19_R2_001.fastq.gz mibi1453_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1454/S_epi_K_1_gross_S20_R1_001.fastq.gz mibi1454_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1454/S_epi_K_1_gross_S20_R2_001.fastq.gz mibi1454_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1455/S_epi_K_2_S21_R1_001.fastq.gz mibi1455_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1455/S_epi_K_2_S21_R2_001.fastq.gz mibi1455_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1456/S_epi_K_3_S22_R1_001.fastq.gz mibi1456_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1456/S_epi_K_3_S22_R2_001.fastq.gz mibi1456_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1457/S_epi_K_6_S23_R1_001.fastq.gz mibi1457_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1457/S_epi_K_6_S23_R2_001.fastq.gz mibi1457_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1458/S_epi_K_8_S24_R1_001.fastq.gz mibi1458_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1458/S_epi_K_8_S24_R2_001.fastq.gz mibi1458_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1459/S_epi_K_17_S25_R1_001.fastq.gz mibi1459_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1459/S_epi_K_17_S25_R2_001.fastq.gz mibi1459_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1460/S_epi_K_26_S26_R1_001.fastq.gz mibi1460_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1460/S_epi_K_26_S26_R2_001.fastq.gz mibi1460_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1461/S_epi_K_34_S27_R1_001.fastq.gz mibi1461_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1461/S_epi_K_34_S27_R2_001.fastq.gz mibi1461_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1462/S_epi_K_35_S28_R1_001.fastq.gz mibi1462_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1462/S_epi_K_35_S28_R2_001.fastq.gz mibi1462_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1463/S_epi_K_19_S29_R1_001.fastq.gz mibi1463_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1463/S_epi_K_19_S29_R2_001.fastq.gz mibi1463_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1464/S_epi_K_22_S30_R1_001.fastq.gz mibi1464_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1464/S_epi_K_22_S30_R2_001.fastq.gz mibi1464_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1465/S_epi_K_23_S31_R1_001.fastq.gz mibi1465_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1465/S_epi_K_23_S31_R2_001.fastq.gz mibi1465_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1466/S_epi_K_24_S32_R1_001.fastq.gz mibi1466_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1466/S_epi_K_24_S32_R2_001.fastq.gz mibi1466_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1467/S_epi_K_30_S33_R1_001.fastq.gz mibi1467_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1467/S_epi_K_30_S33_R2_001.fastq.gz mibi1467_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1468/S_epi_K_31_S34_R1_001.fastq.gz mibi1468_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1468/S_epi_K_31_S34_R2_001.fastq.gz mibi1468_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1469/S_epi_K_32_S35_R1_001.fastq.gz mibi1469_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1469/S_epi_K_32_S35_R2_001.fastq.gz mibi1469_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1470/S_epi_K_1_klein_S36_R1_001.fastq.gz mibi1470_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1470/S_epi_K_1_klein_S36_R2_001.fastq.gz mibi1470_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1471/S_epi_K_37_S37_R1_001.fastq.gz mibi1471_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1471/S_epi_K_37_S37_R2_001.fastq.gz mibi1471_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1472/S_epi_K_46_S38_R1_001.fastq.gz mibi1472_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1472/S_epi_K_46_S38_R2_001.fastq.gz mibi1472_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1473/S_epi_K_57_S39_R1_001.fastq.gz mibi1473_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1473/S_epi_K_57_S39_R2_001.fastq.gz mibi1473_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1474/S_epi_K_59_S40_R1_001.fastq.gz mibi1474_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1474/S_epi_K_59_S40_R2_001.fastq.gz mibi1474_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1475/S_epi_K_58_S41_R1_001.fastq.gz mibi1475_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1475/S_epi_K_58_S41_R2_001.fastq.gz mibi1475_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1476/S_epi_K_60_S42_R1_001.fastq.gz mibi1476_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1476/S_epi_K_60_S42_R2_001.fastq.gz mibi1476_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1477/S_epi_K_61_S43_R1_001.fastq.gz mibi1477_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1477/S_epi_K_61_S43_R2_001.fastq.gz mibi1477_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1478/S_epi_K_28_S44_R1_001.fastq.gz mibi1478_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1478/S_epi_K_28_S44_R2_001.fastq.gz mibi1478_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1479/S_epi_K_33_S45_R1_001.fastq.gz mibi1479_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1479/S_epi_K_33_S45_R2_001.fastq.gz mibi1479_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1480/S_epi_K_39_S46_R1_001.fastq.gz mibi1480_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1480/S_epi_K_39_S46_R2_001.fastq.gz mibi1480_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1481/S_epi_K_40_S47_R1_001.fastq.gz mibi1481_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1481/S_epi_K_40_S47_R2_001.fastq.gz mibi1481_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1482/S_epi_K_43_S48_R1_001.fastq.gz mibi1482_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1482/S_epi_K_43_S48_R2_001.fastq.gz mibi1482_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1483/S_epi_K_97_S49_R1_001.fastq.gz mibi1483_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1483/S_epi_K_97_S49_R2_001.fastq.gz mibi1483_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1484/S_epi_K_84_S50_R1_001.fastq.gz mibi1484_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1484/S_epi_K_84_S50_R2_001.fastq.gz mibi1484_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1485/S_epi_K_85_S51_R1_001.fastq.gz mibi1485_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1485/S_epi_K_85_S51_R2_001.fastq.gz mibi1485_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1486/S_epi_K_86_S52_R1_001.fastq.gz mibi1486_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1486/S_epi_K_86_S52_R2_001.fastq.gz mibi1486_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1487/S_epi_K_116_S53_R1_001.fastq.gz mibi1487_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1487/S_epi_K_116_S53_R2_001.fastq.gz mibi1487_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1488/S_epi_K_118_S54_R1_001.fastq.gz mibi1488_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1488/S_epi_K_118_S54_R2_001.fastq.gz mibi1488_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1489/S_epi_K_120_S55_R1_001.fastq.gz mibi1489_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1489/S_epi_K_120_S55_R2_001.fastq.gz mibi1489_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1490/S_epi_K_123_S56_R1_001.fastq.gz mibi1490_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1490/S_epi_K_123_S56_R2_001.fastq.gz mibi1490_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1491/S_epi_K_124_S57_R1_001.fastq.gz mibi1491_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1491/S_epi_K_124_S57_R2_001.fastq.gz mibi1491_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1492/S_epi_K_127_S58_R1_001.fastq.gz mibi1492_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1492/S_epi_K_127_S58_R2_001.fastq.gz mibi1492_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1493/S_epi_K_29_S59_R1_001.fastq.gz mibi1493_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1493/S_epi_K_29_S59_R2_001.fastq.gz mibi1493_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1494/S_epi_K_72_S60_R1_001.fastq.gz mibi1494_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1494/S_epi_K_72_S60_R2_001.fastq.gz mibi1494_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1495/S_epi_K_89_S61_R1_001.fastq.gz mibi1495_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1495/S_epi_K_89_S61_R2_001.fastq.gz mibi1495_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1496/S_epi_K_110_S62_R1_001.fastq.gz mibi1496_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1496/S_epi_K_110_S62_R2_001.fastq.gz mibi1496_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1497/S_epi_K_112_S63_R1_001.fastq.gz mibi1497_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1497/S_epi_K_112_S63_R2_001.fastq.gz mibi1497_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1498/S_epi_K_114_S64_R1_001.fastq.gz mibi1498_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1498/S_epi_K_114_S64_R2_001.fastq.gz mibi1498_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1499/S_epi_K_91_S65_R1_001.fastq.gz mibi1499_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1499/S_epi_K_91_S65_R2_001.fastq.gz mibi1499_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1500/S_epi_K_92_S66_R1_001.fastq.gz mibi1500_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1500/S_epi_K_92_S66_R2_001.fastq.gz mibi1500_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1501/S_epi_K_95_S67_R1_001.fastq.gz mibi1501_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1501/S_epi_K_95_S67_R2_001.fastq.gz mibi1501_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1502/S_epi_K_96_S68_R1_001.fastq.gz mibi1502_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1502/S_epi_K_96_S68_R2_001.fastq.gz mibi1502_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1503/S_epi_K_98_S69_R1_001.fastq.gz mibi1503_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1503/S_epi_K_98_S69_R2_001.fastq.gz mibi1503_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1504/S_epi_K_100_S70_R1_001.fastq.gz mibi1504_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1504/S_epi_K_100_S70_R2_001.fastq.gz mibi1504_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1505/S_epi_K_106_S71_R1_001.fastq.gz mibi1505_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1505/S_epi_K_106_S71_R2_001.fastq.gz mibi1505_R2.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1506/S_epi_K_108_S72_R1_001.fastq.gz mibi1506_R1.fastq.gz ln -s ../200615_NB501882_0207_AH5YFGAFX2/mibi1506/S_epi_K_108_S72_R2_001.fastq.gz mibi1506_R2.fastq.gz -
start bacto pipeline until step shovill (set true for the first 2 steps: assembly + typing_mlst)
(bengal3_ac3) jhuang@WS-2290C:~/DATA/Data_Luise_Sepi_STKN$ /home/jhuang/miniconda3/envs/snakemake_4_3_1/bin/snakemake --printshellcmds -
concate the first assembly
find . -name "contigs.fa" -exec sh -c 'echo -n "{}: "; wc -c < "{}"; echo " contigs: $(grep -c ">" "{}")"' \; find . -name "contigs.fa" -exec sh -c 'wc -c < "{}" | tr -d "\n" | awk "{print \"{}: \" \$0 \" contigs: \" \$(grep -c \">\" \"{}\")} " ' \; | sort -k2,2n -k4,4n find . -name "contigs.fa" -exec sh -c 'size=$(wc -c < "{}"); contig_count=$(grep -c ">" "{}"); printf "%s: %d bytes, contigs: %d\n" "{}" "$size" "$contig_count"' \; | sort -t: -k4,4n grep -v '^>' contigs.fa | awk '{ total += length($0) } END { print total }' #2399339 find . -name "contigs.fa" -exec sh -c 'total_length=$(grep -v "^>" "{}" | awk "{ total += length(\$0) } END { print total }"); printf "%s: %d\n" "{}" "$total_length"' \; | sort -t: -k2,2n ./mibi2381/contigs.fa: 1077606 ./mibi1472/contigs.fa: 1145583 ./mibi2380/contigs.fa: 1478149 ./mibi1458/contigs.fa: 2345604 ./mibi1466/contigs.fa: 2349339 ./mibi1451/contigs.fa: 2375432 ./mibi1441/contigs.fa: 2390897 ./mibi1471/contigs.fa: 2399056 ./mibi1435/contigs.fa: 2399339 ./mibi1473/contigs.fa: 2416173 ./mibi1474/contigs.fa: 2419849 ./mibi1495/contigs.fa: 2423463 ./mibi1497/contigs.fa: 2427317 ./mibi1476/contigs.fa: 2428189 ./mibi1463/contigs.fa: 2430568 ./mibi2313/contigs.fa: 2434775 ./mibi2321/contigs.fa: 2435584 ./mibi1494/contigs.fa: 2436708 ./mibi1481/contigs.fa: 2437976 ./mibi1464/contigs.fa: 2438603 ./mibi1450/contigs.fa: 2439465 ./mibi1483/contigs.fa: 2440401 ./mibi1506/contigs.fa: 2440413 ./mibi1462/contigs.fa: 2441903 ./mibi1443/contigs.fa: 2445186 ./mibi1448/contigs.fa: 2447751 ./mibi1439/contigs.fa: 2453223 ./mibi1501/contigs.fa: 2454766 ./mibi1505/contigs.fa: 2455113 ./mibi1436/contigs.fa: 2457498 ./mibi1499/contigs.fa: 2459216 ./mibi1452/contigs.fa: 2467292 ./mibi1500/contigs.fa: 2467976 ./mibi1455/contigs.fa: 2468334 ./mibi1445/contigs.fa: 2469433 ./mibi1479/contigs.fa: 2471443 ./mibi1482/contigs.fa: 2473012 ./mibi1469/contigs.fa: 2475757 ./mibi1468/contigs.fa: 2478455 ./mibi1438/contigs.fa: 2482554 ./mibi2379/contigs.fa: 2483292 ./mibi1493/contigs.fa: 2483445 ./mibi1488/contigs.fa: 2484950 ./mibi1486/contigs.fa: 2485023 ./mibi1442/contigs.fa: 2488933 ./mibi1437/contigs.fa: 2489787 ./mibi1460/contigs.fa: 2491014 ./mibi2312/contigs.fa: 2494588 ./mibi1489/contigs.fa: 2495771 ./mibi1504/contigs.fa: 2501045 ./mibi2317/contigs.fa: 2504746 ./mibi1444/contigs.fa: 2504874 ./mibi1446/contigs.fa: 2508952 ./mibi1465/contigs.fa: 2509542 ./mibi1467/contigs.fa: 2509571 ./mibi2314/contigs.fa: 2510798 ./mibi2315/contigs.fa: 2511464 ./mibi1457/contigs.fa: 2513005 ./mibi1490/contigs.fa: 2514271 ./mibi1491/contigs.fa: 2519558 ./mibi1449/contigs.fa: 2520352 ./mibi1503/contigs.fa: 2521179 ./mibi1475/contigs.fa: 2526469 ./mibi1484/contigs.fa: 2530724 ./mibi1447/contigs.fa: 2538610 ./mibi1453/contigs.fa: 2543893 ./mibi1477/contigs.fa: 2550922 ./mibi1454/contigs.fa: 2553326 ./mibi1498/contigs.fa: 2556315 ./mibi2316/contigs.fa: 2556628 ./mibi1492/contigs.fa: 2558492 ./mibi1470/contigs.fa: 2563169 ./mibi2320/contigs.fa: 2571421 ./mibi1461/contigs.fa: 2582715 ./mibi1478/contigs.fa: 2584524 ./mibi2318/contigs.fa: 2594120 ./mibi1459/contigs.fa: 2595990 ./mibi1440/contigs.fa: 2604213 ./mibi1456/contigs.fa: 2605647 ./mibi1487/contigs.fa: 2614950 ./mibi1496/contigs.fa: 2616533 ./mibi2319/contigs.fa: 2621258 ./mibi1480/contigs.fa: 2621312 ./mibi1485/contigs.fa: 2665904 ./mibi1502/contigs.fa: 2668891 cd mibi1502 awk '/^>/ {if(seen_header++) print "NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN"; next} {printf "%s", $0} END {print ""}' contigs.fa > concatenated_contigs.fasta awk '/^>/ {if(seen_header++) print ""; print $0; next} {printf "%s", $0} END {print ""}' concatenated_contigs.fasta > concatenated_contigs.fa sed -i '1s/^/>mibi1502\n/' concatenated_contigs.fa seqkit seq -i concatenated_contigs.fa -m 100 > ../mibi1502.fasta bakta --db /mnt/nvme0n1p1/bakta_db mibi1502.fasta ATCC 14990 Genes (total) :: 2,368 CDSs (total) :: 2,284 Genes (coding) :: 2,231 CDSs (with protein) :: 2,231 Genes (RNA) :: 84 rRNAs :: 7, 6, 6 (5S, 16S, 23S) complete rRNAs :: 7, 6, 6 (5S, 16S, 23S) tRNAs :: 61 ncRNAs :: 4 Pseudo Genes (total) :: 53 CDSs (without protein) :: 53 Pseudo Genes (ambiguous residues) :: 0 of 53 Pseudo Genes (frameshifted) :: 34 of 53 Pseudo Genes (incomplete) :: 31 of 53 Pseudo Genes (internal stop) :: 24 of 53 Detected IPSs: 2369 This likely refers to insertion sequences or insertional plasmids (depending on the specific context). In this case, 2,369 insertion sequences were detected in the analysis, which could indicate mobile genetic elements that might affect the genome's stability or function. predict tRNAs... found: 61 predict tmRNAs... found: 1 predict rRNAs... found: 12 predict ncRNAs... found: 55 predict ncRNA regions... found: 26 predict CRISPR arrays... found: 0 predict & annotate CDSs... predicted: 2529 discarded spurious: 0 revised translational exceptions: 0 detected IPSs: 2369 found PSCs: 117 found PSCCs: 26 lookup annotations... conduct expert systems... amrfinder: 20 protein sequences: 54 combine annotations and mark hypotheticals... detect pseudogenes... pseudogene candidates: 15 found pseudogenes: 7 analyze hypothetical proteins: 96 detected Pfam hits: 1 calculated proteins statistics revise special cases... extract sORF... potential: 46973 discarded due to overlaps: 37990 discarded spurious: 0 detected IPSs: 9 found PSCs: 2 lookup annotations... filter and combine annotations... filtered sORFs: 9 detect gaps... found: 113 detect oriCs/oriVs... found: 1 detect oriTs... found: 0 apply feature overlap filters... select features and create locus tags... selected: 2801 improve annotations... revised gene symbols: 9 -
generate genebank in snpEff and run spandx
#mkdir ~/miniconda3/envs/spandx/share/snpeff-5.1-2/data/NZ_CP035288 #cp NZ_CP035288.gb ~/miniconda3/envs/spandx/share/snpeff-5.1-2/data/NZ_CP035288/genes.gbk #vim ~/miniconda3/envs/spandx/share/snpeff-5.1-2/snpEff.config #/home/jhuang/miniconda3/envs/spandx/bin/snpEff build -genbank NZ_CP035288 #-d cp mibi1502.gbff ../db/mibi1502.gb #Then replace contig_1 to mibi1502 in the gb-file cp mibi1502.fasta ../db/mibi1502.fasta mkdir ~/miniconda3/envs/spandx/share/snpeff-5.1-2/data/mibi1502 cp mibi1502.gb ~/miniconda3/envs/spandx/share/snpeff-5.1-2/data/mibi1502/genes.gbk vim ~/miniconda3/envs/spandx/share/snpeff-5.1-2/snpEff.config #Protein check: mibi1502 OK: 2525 Not found: 7 Errors: 0 Error percentage: 0.0% /home/jhuang/miniconda3/envs/spandx/bin/snpEff build -genbank mibi1502 #-d # run the first 5 steps in bacto-pipeline ("pangenome"+"variants_calling"+"phylogeny_fasttree"+"phylogeny_raxml"+"recombination") /home/jhuang/miniconda3/envs/snakemake_4_3_1/bin/snakemake --printshellcmds ln -s /home/jhuang/Tools/spandx/ spandx # cp trimmed trimmed_gzipped; cd trimmed_gzipped; gzip *.fastq; cd .. (spandx) nextflow run spandx/main.nf --fastq "trimmed_gzipped/*_P_{1,2}.fastq.gz" --ref db/mibi1502.fasta --annotation --database mibi1502 -resume mv work work_mibi1502 mv Outputs Outputs_mibi1502 -
filter out 4 samples for downstream analyses
#The coverages of shovill/mibi1470/contigs.fa and shovill/mibi1472/contigs.fa, and shovill/mibi2381/contigs.fa are low. #mibi1470 could not be identified as S. epidermidis based on its MLST type. #remove: mibi1470 2.5M #remove: ./mibi1472/contigs.fa: 1145583 #remove: ./mibi2380/contigs.fa: 1478149 #remove: ./mibi2381/contigs.fa: 1077606 #Then we have 81 samples for downstream analysis: mibi1435 -- mibi1506 (72); mibi2312 -- mibi2321 (10); mibi2379 -- mibi2381 (3) -
SCCmec typing and drawing with clinker
#https://www.genomicepidemiology.org/ #https://cge.food.dtu.dk/services/SCCmecFinder/ mkdir contigs; cd contigs for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do cp ../${sample}/contigs.fa ${sample}_contigs.fa done #Predicted SCCmec element grep "Predicted SCCmec element" *.txt grep "2 SCCmec" *.txt grep "ccr class" *.txt grep "mec class" *.txt mibi1435_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IVb(2B) mibi1436_SCCmec_out.txt:Predicted SCCmec element: none mibi1437_SCCmec_out.txt:Predicted SCCmec element: none mibi1438_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IV(2B) mibi1439_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IVa(2B) mibi1440_SCCmec_out.txt:Predicted SCCmec element: none mibi1441_SCCmec_out.txt:Predicted SCCmec element: none mibi1442_SCCmec_out.txt:Predicted SCCmec element: none mibi1443_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IVg(2B) mibi1444_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IV(2B&5) mibi1445_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IVa(2B) mibi1446_SCCmec_out.txt: SCCmec_type_IV(2B) and SCCmec_type_VI(4B) mibi1447_SCCmec_out.txt: SCCmec_type_IV(2B) and SCCmec_type_VI(4B) mibi1448_SCCmec_out.txt:Predicted SCCmec element: none mibi1449_SCCmec_out.txt:Predicted SCCmec element: none mibi1450_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IV(2B) mibi1451_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IV(2B) mibi1452_SCCmec_out.txt:Predicted SCCmec element: none mibi1453_SCCmec_out.txt:Predicted SCCmec element: none mibi1454_SCCmec_out.txt:Predicted SCCmec element: none mibi1455_SCCmec_out.txt:Predicted SCCmec element: none mibi1456_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_III(3A) mibi1457_SCCmec_out.txt: SCCmec_type_IV(2B) and SCCmec_type_VI(4B) mibi1458_SCCmec_out.txt:Predicted SCCmec element: none mibi1459_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_III(3A) mibi1460_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IVa(2B) mibi1461_SCCmec_out.txt:Predicted SCCmec element: none mibi1462_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IVg(2B) mibi1463_SCCmec_out.txt:Predicted SCCmec element: none mibi1464_SCCmec_out.txt:Predicted SCCmec element: none mibi1465_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IV(2B&5) mibi1466_SCCmec_out.txt: SCCmec_type_IV(2B) and SCCmec_type_VI(4B) mibi1467_SCCmec_out.txt:Predicted SCCmec element: none mibi1468_SCCmec_out.txt:Predicted SCCmec element: none mibi1469_SCCmec_out.txt:Predicted SCCmec element: none mibi1471_SCCmec_out.txt:Predicted SCCmec element: none mibi1473_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IVa(2B) mibi1474_SCCmec_out.txt:Predicted SCCmec element: none mibi1475_SCCmec_out.txt: SCCmec_type_IV(2B) and SCCmec_type_VI(4B) mibi1476_SCCmec_out.txt:Predicted SCCmec element: none mibi1477_SCCmec_out.txt:Predicted SCCmec element: none mibi1478_SCCmec_out.txt: SCCmec_type_III(3A) and SCCmec_type_VIII(4A) mibi1479_SCCmec_out.txt: SCCmec_type_IV(2B) and SCCmec_type_VI(4B) mibi1480_SCCmec_out.txt: SCCmec_type_III(3A) and SCCmec_type_VIII(4A) mibi1481_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_II(2A) mibi1482_SCCmec_out.txt:Predicted SCCmec element: none mibi1483_SCCmec_out.txt:Predicted SCCmec element: none mibi1484_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IV(2B&5) mibi1485_SCCmec_out.txt:Predicted SCCmec element: none mibi1486_SCCmec_out.txt:Predicted SCCmec element: none mibi1487_SCCmec_out.txt: SCCmec_type_III(3A) and SCCmec_type_VIII(4A) mibi1488_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IV(2B) mibi1489_SCCmec_out.txt:Predicted SCCmec element: none mibi1490_SCCmec_out.txt: SCCmec_type_IV(2B) and SCCmec_type_VI(4B) mibi1491_SCCmec_out.txt: SCCmec_type_IV(2B) and SCCmec_type_VI(4B) mibi1492_SCCmec_out.txt:Predicted SCCmec element: none mibi1493_SCCmec_out.txt:Predicted SCCmec element: none mibi1494_SCCmec_out.txt:Predicted SCCmec element: none mibi1495_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IVa(2B) mibi1496_SCCmec_out.txt:Predicted SCCmec element: none mibi1497_SCCmec_out.txt:Predicted SCCmec element: none mibi1498_SCCmec_out.txt:Predicted SCCmec element: none mibi1499_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IVa(2B) mibi1500_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IVa(2B) mibi1501_SCCmec_out.txt: SCCmec_type_IV(2B) and SCCmec_type_VI(4B) mibi1502_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_III(3A) mibi1503_SCCmec_out.txt:Predicted SCCmec element: none mibi1504_SCCmec_out.txt:Predicted SCCmec element: none mibi1505_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IV(2B&5) mibi1506_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IVg(2B) mibi2312_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IV(2B&5) mibi2313_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IV(2B) mibi2314_SCCmec_out.txt:Predicted SCCmec element: none mibi2315_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IVa(2B) mibi2316_SCCmec_out.txt:Predicted SCCmec element: none mibi2317_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IV(2B&5) mibi2318_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_III(3A) mibi2319_SCCmec_out.txt: SCCmec_type_III(3A) and SCCmec_type_VIII(4A) mibi2320_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IV(2B&5) mibi2321_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IV(2B) mibi2379_SCCmec_out.txt:Predicted SCCmec element: SCCmec_type_IV(2B&5) mibi1441_SCCmec_out.txt:ccr class 2 mibi1446_SCCmec_out.txt:ccr class 2 mibi1446_SCCmec_out.txt:ccr class 4 mibi1447_SCCmec_out.txt:ccr class 2 mibi1447_SCCmec_out.txt:ccr class 4 mibi1452_SCCmec_out.txt:ccr class 5 mibi1455_SCCmec_out.txt:ccr class 2 mibi1457_SCCmec_out.txt:ccr class 2 mibi1457_SCCmec_out.txt:ccr class 4 mibi1464_SCCmec_out.txt:ccr class 2 mibi1466_SCCmec_out.txt:ccr class 2 mibi1466_SCCmec_out.txt:ccr class 4 mibi1467_SCCmec_out.txt:ccr class 5 mibi1475_SCCmec_out.txt:ccr class 2 mibi1475_SCCmec_out.txt:ccr class 4 mibi1477_SCCmec_out.txt:ccr class 9 mibi1478_SCCmec_out.txt:ccr class 3 mibi1478_SCCmec_out.txt:ccr class 4 mibi1479_SCCmec_out.txt:ccr class 2 mibi1479_SCCmec_out.txt:ccr class 4 mibi1480_SCCmec_out.txt:ccr class 3 mibi1480_SCCmec_out.txt:ccr class 4 mibi1482_SCCmec_out.txt:ccr class 5 mibi1483_SCCmec_out.txt:ccr class 5 mibi1486_SCCmec_out.txt:ccr class 2 mibi1487_SCCmec_out.txt:ccr class 3 mibi1487_SCCmec_out.txt:ccr class 4 mibi1489_SCCmec_out.txt:ccr class 2 mibi1490_SCCmec_out.txt:ccr class 2 mibi1490_SCCmec_out.txt:ccr class 4 mibi1491_SCCmec_out.txt:ccr class 2 mibi1491_SCCmec_out.txt:ccr class 4 mibi1492_SCCmec_out.txt:ccr class 2 mibi1494_SCCmec_out.txt:ccr class 5 mibi1501_SCCmec_out.txt:ccr class 2 mibi1501_SCCmec_out.txt:ccr class 4 mibi1504_SCCmec_out.txt:ccr class 2 mibi1504_SCCmec_out.txt:ccr class 5&5 mibi2316_SCCmec_out.txt:ccr class 2 mibi2319_SCCmec_out.txt:ccr class 3 mibi2319_SCCmec_out.txt:ccr class 4 mibi1446_SCCmec_out.txt:mec class B mibi1447_SCCmec_out.txt:mec class B mibi1457_SCCmec_out.txt:mec class B mibi1463_SCCmec_out.txt:mec class B mibi1466_SCCmec_out.txt:mec class B mibi1475_SCCmec_out.txt:mec class B mibi1478_SCCmec_out.txt:mec class A mibi1479_SCCmec_out.txt:mec class B mibi1480_SCCmec_out.txt:mec class A mibi1483_SCCmec_out.txt:mec class A mibi1487_SCCmec_out.txt:mec class A mibi1490_SCCmec_out.txt:mec class B mibi1491_SCCmec_out.txt:mec class B mibi1501_SCCmec_out.txt:mec class B mibi2319_SCCmec_out.txt:mec class A -
agr typing
for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do bakta --db /mnt/nvme0n1p1/bakta_db shovill/${sample}/contigs.fa --prefix ${sample} done grep "agrD" mibi1436.gbff mibi1435 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1436 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1437 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1438 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1439 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1440 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1441 MNLLGGLLLKLFSNFMAVIGNAAKYNPCASYLDEPQVPEELTKLDE AgrD III mibi1442 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1443 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1444 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1445 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1446 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1447 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1448 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1449 MNLLGGLLLKLFSNFMAVIGNAAKYNPCASYLDEPQVPEELTKLDE AgrD III mibi1450 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1451 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1452 MNLLGGLLLKLFSNFMAVIGNAAKYNPCASYLDEPQVPEELTKLDE AgrD III mibi1453 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1454 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1455 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1456 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1457 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1458 MENIFNLFIK I FTTILEF V GTVAGDSVCASYFDEPEVPEELTKLYE AgrD I* (with a 2-codon difference) mibi1459 MENIFNLFIK F FTTILEF I GTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1460 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1461 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1462 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1463 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1464 MNLLGGLLLKLFSNFMAVIGNAAKYNPCASYLDEPQVPEELTKLDE AgrD III mibi1465 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1466 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1467 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1468 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1469 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1471 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1473 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1474 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1475 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1476 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1477 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1478 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1479 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1480 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1481 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1482 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1483 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1484 none mibi1485 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1486 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1487 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1488 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1489 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1490 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1491 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1492 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1493 MNLLGGL F LK I FSNFMAVIGNA A KYNPC AS YLDEPQVPEELTKLDE AgrD III* (with a 2-codon difference) mibi1494 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1495 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1496 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1497 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1498 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1499 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi1500 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1501 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1502 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1503 MNLLGGLLLKLFSNFMAVIGNAAKYNPCASYLDEPQVPEELTKLDE AgrD III mibi1504 MNLLGGLLLKLFSNFMAVIGNAAKYNPCASYLDEPQVPEELTKLDE AgrD III mibi1505 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi1506 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi2312 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi2313 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi2314 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi2315 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi2316 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi2317 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi2318 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi2319 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi2320 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II mibi2321 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE AgrD I mibi2379 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE AgrD II #MNLLGGL L LK L FSNFMAVIGNA A KYNPC AS YLDEPQVPEELTKLDE AgrD III (2) #MNLLGGL L LK I FSNFMAVIGNA S KYNPC SN YLDEPQVPEELTKLDE AgrD II (4) -- AgrD I -- Query 1 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE 46 M + L +K F+ + IG + + C Y DEP+VPEELTKL E Sbjct 926825 MNLLGGLLLKIFSNFMAVIGNASKYNPCVMYLDEPQVPEELTKLDE 926688 -- AgrD II -- Query 1 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE 46 MNLLGGLLLKIFSNFMAVIGNASKYNPC YLDEPQVPEELTKLDE Sbjct 926825 MNLLGGLLLKIFSNFMAVIGNASKYNPCVMYLDEPQVPEELTKLDE 926688 -- AgrD III -- Query 1 MNLLGGLLLKLFSNFMAVIGNAAKYNPCASYLDEPQVPEELTKLDE 46 MNLLGGLLLK+FSNFMAVIGNA+KYNPC YLDEPQVPEELTKLDE Sbjct 926825 MNLLGGLLLKIFSNFMAVIGNASKYNPCVMYLDEPQVPEELTKLDE 926688 -
Calulate the presence-absence-matrix by searching keyword gene=gene_name in gff3-files
cut -d',' -f1 plotTreeHeatmap/typing.csv > all_isolate_names.txt grep "gene=gyrB" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 grep "gene=fumC" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 grep "gene=gltA" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 grep "gene=icd" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 grep "gene=apsS" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 | wc -l #TODO, using sequence comparisons for one record! grep "gene=sigB" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 #27d26 #< mibi1461 grep "gene=sarA" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 #17d16 #< mibi1451 #32d30 #< mibi1466 #45d42 #< mibi1481 grep "gene=agrC" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 | wc -l #TODO, using sequence comparisons for one record! grep "gene=yycG" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 | wc -l #TODO, using sequence comparisons for one record! grep "PSM-beta" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 | wc -l #TODO, using sequence comparisons for one record! grep "PSM-delta" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 #9d8 #< mibi1443 grep "hlb" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 | wc -l #TODO, using sequence comparisons for one record! grep "atlE" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 | wc -l #TODO, using sequence comparisons for one record! grep "gene=atl" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 #19d18 #< mibi1453 grep "gene=sdrG" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 #2d1 < mibi1436 5d3 < mibi1439 8,9d5 < mibi1442 < mibi1443 11,12d6 < mibi1445 < mibi1446 14d7 < mibi1448 18,19d10 < mibi1452 < mibi1453 30,32d20 < mibi1464 < mibi1465 < mibi1466 34,35d21 < mibi1468 < mibi1469 37d22 < mibi1473 39d23 < mibi1475 41d24 < mibi1477 43d25 < mibi1479 47d28 < mibi1483 50d30 < mibi1486 52d31 < mibi1488 55d33 < mibi1491 63,64d40 < mibi1499 < mibi1500 67d42 < mibi1503 76d50 < mibi2317 79d52 < mibi2320 81d53 < mibi2379 grep "gene=sdrH" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 #24d23 #< mibi1458 #57d55 #< mibi1493 grep "gene=ebh" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 #1d0 #< mibi1435 6d4 < mibi1440 11,13d8 < mibi1445 < mibi1446 < mibi1447 16,18d10 < mibi1450 < mibi1451 < mibi1452 20d11 < mibi1454 22,23d12 < mibi1456 < mibi1457 25,27d13 < mibi1459 < mibi1460 < mibi1461 29d14 < mibi1463 31,32d15 < mibi1465 < mibi1466 37d19 < mibi1473 39d20 < mibi1475 41,44d21 < mibi1477 < mibi1478 < mibi1479 < mibi1480 47d23 < mibi1483 49d24 < mibi1485 51,52d25 < mibi1487 < mibi1488 54,55d26 < mibi1490 < mibi1491 59,60d29 < mibi1495 < mibi1496 62,63d30 < mibi1498 < mibi1499 66d32 < mibi1502 71d36 < mibi2312 74d38 < mibi2315 76,79d39 < mibi2317 < mibi2318 < mibi2319 < mibi2320 81d40 < mibi2379 grep "gene=ebpS" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 grep "gene=tagB" *.gff3 > temp cut -d'.' -f1 temp > temp1 uniq temp1 > temp2 # 3 copies for each isolate diff all_isolate_names.txt temp2 grep "gene=capC" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 | wc -l #TODO, using sequence comparisons for one record! grep "gene=sepA" *.gff3 > temp cut -d'.' -f1 temp > temp1 uniq temp1 > temp2 diff all_isolate_names.txt temp2 grep "gene=dltA" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 grep "gene=fmtC" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 | wc -l #TODO, using sequence comparisons for one record! grep "gene=lipA" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 grep "gene=sceD" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 #TODO: check the sdrG more detailed with sequence alignments, since SE0760 is not standard gene name. grep "gene=SE0760" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 | wc -l #TODO, using sequence comparisons for one record! grep "gene=esp" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 | wc -l #TODO, using sequence comparisons for one record! grep "gene=ecpA" *.gff3 > temp cut -d'.' -f1 temp > temp1 diff all_isolate_names.txt temp1 17d16 < mibi1451 37d35 < mibi1473 -
Calulate the presence-absence-matrix by local blastn searching
(Optional online search) https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&BLAST_SPEC=MicrobialGenomes #Title:Refseq prokaryote representative genomes (contains refseq assembly) #Molecule Type:mixed DNA #Update date:2024/10/16 #Number of sequences:1038672 #cat gyrB_revcomp.fasta fumC_revcomp.fasta gltA.fasta icd_revcomp.fasta apsS.fasta sigB_revcomp.fasta sarA_revcomp.fasta ... cp ~/DATA/Data_Patricia_Sepi_5Samples/presence_absence_matrix_on_gene_list/apsS.fasta . cp ~/DATA/Data_Patricia_Sepi_5Samples/presence_absence_matrix_on_gene_list/agrC.fasta . cp ~/DATA/Data_Patricia_Sepi_5Samples/presence_absence_matrix_on_gene_list/yycG.fasta . cp ~/DATA/Data_Patricia_Sepi_5Samples/presence_absence_matrix_on_gene_list/psm-beta.fasta . cp ~/DATA/Data_Patricia_Sepi_5Samples/presence_absence_matrix_on_gene_list/hlb_.fasta . cp ~/DATA/Data_Patricia_Sepi_5Samples/presence_absence_matrix_on_gene_list/atlE.fasta . cp ~/DATA/Data_Patricia_Sepi_5Samples/presence_absence_matrix_on_gene_list/capC.fasta . cp ~/DATA/Data_Patricia_Sepi_5Samples/presence_absence_matrix_on_gene_list/fmtC.fasta . cp ~/DATA/Data_Patricia_Sepi_5Samples/presence_absence_matrix_on_gene_list/SE0760.fasta . cp ~/DATA/Data_Patricia_Sepi_5Samples/presence_absence_matrix_on_gene_list/esp.fasta . cp ~/DATA/Data_Patricia_Sepi_5Samples/presence_absence_matrix_on_gene_list/MT880870.fasta . cp ~/DATA/Data_Patricia_Sepi_5Samples/presence_absence_matrix_on_gene_list/MT880871.fasta . cp ~/DATA/Data_Patricia_Sepi_5Samples/presence_absence_matrix_on_gene_list/MT880872.fasta . for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do cd ../shovill/${sample}/ makeblastdb -in contigs.fa -dbtype nucl cd ../../presence_absence_matrix_on_gene_list done # -length(apsS.fasta)=1041 for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query apsS.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > apsS_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py apsS_on_${sample}.blastn done apsS_on_mibi1435.blastn: 1041 apsS_on_mibi1436.blastn: 1041 apsS_on_mibi1437.blastn: 1041 apsS_on_mibi1438.blastn: 1041 apsS_on_mibi1439.blastn: 1041 apsS_on_mibi1440.blastn: 1041 apsS_on_mibi1441.blastn: 1041 apsS_on_mibi1442.blastn: 1041 apsS_on_mibi1443.blastn: 1041 apsS_on_mibi1444.blastn: 1041 apsS_on_mibi1445.blastn: 1041 apsS_on_mibi1446.blastn: 1041 apsS_on_mibi1447.blastn: 1041 apsS_on_mibi1448.blastn: 1041 apsS_on_mibi1449.blastn: 1041 apsS_on_mibi1450.blastn: 1041 apsS_on_mibi1451.blastn: 1041 apsS_on_mibi1452.blastn: 1041 apsS_on_mibi1453.blastn: 1041 apsS_on_mibi1454.blastn: 1041 apsS_on_mibi1455.blastn: 1041 apsS_on_mibi1456.blastn: 1041 apsS_on_mibi1457.blastn: 1041 apsS_on_mibi1458.blastn: 1041 apsS_on_mibi1459.blastn: 1041 apsS_on_mibi1460.blastn: 1041 apsS_on_mibi1461.blastn: 1041 apsS_on_mibi1462.blastn: 1041 apsS_on_mibi1463.blastn: 1041 apsS_on_mibi1464.blastn: 1041 apsS_on_mibi1465.blastn: 1041 apsS_on_mibi1466.blastn: 1041 apsS_on_mibi1467.blastn: 1041 apsS_on_mibi1468.blastn: 1041 apsS_on_mibi1469.blastn: 1041 apsS_on_mibi1471.blastn: 1041 apsS_on_mibi1473.blastn: 1041 apsS_on_mibi1474.blastn: 1041 apsS_on_mibi1475.blastn: 1041 apsS_on_mibi1476.blastn: 1041 apsS_on_mibi1477.blastn: 1041 apsS_on_mibi1478.blastn: 1041 apsS_on_mibi1479.blastn: 1041 apsS_on_mibi1480.blastn: 1041 apsS_on_mibi1481.blastn: 1041 apsS_on_mibi1482.blastn: 1041 apsS_on_mibi1483.blastn: 1041 apsS_on_mibi1484.blastn: 1041 apsS_on_mibi1485.blastn: 1041 apsS_on_mibi1486.blastn: 1041 apsS_on_mibi1487.blastn: 1041 apsS_on_mibi1488.blastn: 1041 apsS_on_mibi1489.blastn: 1041 apsS_on_mibi1490.blastn: 1041 apsS_on_mibi1491.blastn: 1041 apsS_on_mibi1492.blastn: 1041 apsS_on_mibi1493.blastn: 1041 apsS_on_mibi1494.blastn: 1041 apsS_on_mibi1495.blastn: 1041 apsS_on_mibi1496.blastn: 1041 apsS_on_mibi1497.blastn: 1041 apsS_on_mibi1498.blastn: 1041 apsS_on_mibi1499.blastn: 1041 apsS_on_mibi1500.blastn: 1041 apsS_on_mibi1501.blastn: 1041 apsS_on_mibi1502.blastn: 1041 apsS_on_mibi1503.blastn: 1041 apsS_on_mibi1504.blastn: 1041 apsS_on_mibi1505.blastn: 1041 apsS_on_mibi1506.blastn: 1041 apsS_on_mibi2312.blastn: 1041 apsS_on_mibi2313.blastn: 1041 apsS_on_mibi2314.blastn: 1041 apsS_on_mibi2315.blastn: 1041 apsS_on_mibi2316.blastn: 1041 apsS_on_mibi2317.blastn: 1041 apsS_on_mibi2318.blastn: 1041 apsS_on_mibi2319.blastn: 1041 apsS_on_mibi2320.blastn: 1041 apsS_on_mibi2321.blastn: 1041 apsS_on_mibi2379.blastn: 1041 # -length(sigB_revcomp.fasta)=771 for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query sigB_revcomp.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sigB_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py sigB_on_${sample}.blastn done sigB_on_mibi1435.blastn: 771 sigB_on_mibi1436.blastn: 771 sigB_on_mibi1437.blastn: 771 sigB_on_mibi1438.blastn: 771 sigB_on_mibi1439.blastn: 771 sigB_on_mibi1440.blastn: 771 sigB_on_mibi1441.blastn: 771 sigB_on_mibi1442.blastn: 771 sigB_on_mibi1443.blastn: 771 sigB_on_mibi1444.blastn: 771 sigB_on_mibi1445.blastn: 771 sigB_on_mibi1446.blastn: 771 sigB_on_mibi1447.blastn: 771 sigB_on_mibi1448.blastn: 771 sigB_on_mibi1449.blastn: 771 sigB_on_mibi1450.blastn: 771 sigB_on_mibi1451.blastn: 771 sigB_on_mibi1452.blastn: 771 sigB_on_mibi1453.blastn: 771 sigB_on_mibi1454.blastn: 771 sigB_on_mibi1455.blastn: 771 sigB_on_mibi1456.blastn: 771 sigB_on_mibi1457.blastn: 771 sigB_on_mibi1458.blastn: 771 sigB_on_mibi1459.blastn: 771 sigB_on_mibi1460.blastn: 771 sigB_on_mibi1461.blastn: 0 * sigB_on_mibi1462.blastn: 771 sigB_on_mibi1463.blastn: 771 sigB_on_mibi1464.blastn: 771 sigB_on_mibi1465.blastn: 771 sigB_on_mibi1466.blastn: 771 sigB_on_mibi1467.blastn: 771 sigB_on_mibi1468.blastn: 771 sigB_on_mibi1469.blastn: 771 sigB_on_mibi1471.blastn: 771 sigB_on_mibi1473.blastn: 771 sigB_on_mibi1474.blastn: 771 sigB_on_mibi1475.blastn: 771 sigB_on_mibi1476.blastn: 771 sigB_on_mibi1477.blastn: 771 sigB_on_mibi1478.blastn: 771 sigB_on_mibi1479.blastn: 771 sigB_on_mibi1480.blastn: 771 sigB_on_mibi1481.blastn: 771 sigB_on_mibi1482.blastn: 771 sigB_on_mibi1483.blastn: 771 sigB_on_mibi1484.blastn: 771 sigB_on_mibi1485.blastn: 771 sigB_on_mibi1486.blastn: 771 sigB_on_mibi1487.blastn: 771 sigB_on_mibi1488.blastn: 771 sigB_on_mibi1489.blastn: 771 sigB_on_mibi1490.blastn: 771 sigB_on_mibi1491.blastn: 771 sigB_on_mibi1492.blastn: 771 sigB_on_mibi1493.blastn: 771 sigB_on_mibi1494.blastn: 771 sigB_on_mibi1495.blastn: 771 sigB_on_mibi1496.blastn: 771 sigB_on_mibi1497.blastn: 771 sigB_on_mibi1498.blastn: 771 sigB_on_mibi1499.blastn: 771 sigB_on_mibi1500.blastn: 771 sigB_on_mibi1501.blastn: 771 sigB_on_mibi1502.blastn: 771 sigB_on_mibi1503.blastn: 771 sigB_on_mibi1504.blastn: 771 sigB_on_mibi1505.blastn: 771 sigB_on_mibi1506.blastn: 771 sigB_on_mibi2312.blastn: 771 sigB_on_mibi2313.blastn: 771 sigB_on_mibi2314.blastn: 771 sigB_on_mibi2315.blastn: 771 sigB_on_mibi2316.blastn: 771 sigB_on_mibi2317.blastn: 771 sigB_on_mibi2318.blastn: 771 sigB_on_mibi2319.blastn: 771 sigB_on_mibi2320.blastn: 771 sigB_on_mibi2321.blastn: 771 sigB_on_mibi2379.blastn: 771 # - length(sarA_revcomp.fasta)=375 for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query sarA_revcomp.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sarA_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py sarA_on_${sample}.blastn done sarA_on_mibi1435.blastn: 375 sarA_on_mibi1436.blastn: 375 sarA_on_mibi1437.blastn: 375 sarA_on_mibi1438.blastn: 375 sarA_on_mibi1439.blastn: 375 sarA_on_mibi1440.blastn: 375 sarA_on_mibi1441.blastn: 375 sarA_on_mibi1442.blastn: 375 sarA_on_mibi1443.blastn: 375 sarA_on_mibi1444.blastn: 375 sarA_on_mibi1445.blastn: 375 sarA_on_mibi1446.blastn: 375 sarA_on_mibi1447.blastn: 375 sarA_on_mibi1448.blastn: 375 sarA_on_mibi1449.blastn: 375 sarA_on_mibi1450.blastn: 375 sarA_on_mibi1451.blastn: 0 * sarA_on_mibi1452.blastn: 375 sarA_on_mibi1453.blastn: 375 sarA_on_mibi1454.blastn: 375 sarA_on_mibi1455.blastn: 375 sarA_on_mibi1456.blastn: 375 sarA_on_mibi1457.blastn: 375 sarA_on_mibi1458.blastn: 375 sarA_on_mibi1459.blastn: 375 sarA_on_mibi1460.blastn: 375 sarA_on_mibi1461.blastn: 375 sarA_on_mibi1462.blastn: 375 sarA_on_mibi1463.blastn: 375 sarA_on_mibi1464.blastn: 375 sarA_on_mibi1465.blastn: 375 sarA_on_mibi1466.blastn: 208 * sarA_on_mibi1467.blastn: 375 sarA_on_mibi1468.blastn: 375 sarA_on_mibi1469.blastn: 375 sarA_on_mibi1471.blastn: 375 sarA_on_mibi1473.blastn: 375 sarA_on_mibi1474.blastn: 375 sarA_on_mibi1475.blastn: 375 sarA_on_mibi1476.blastn: 375 sarA_on_mibi1477.blastn: 375 sarA_on_mibi1478.blastn: 375 sarA_on_mibi1479.blastn: 375 sarA_on_mibi1480.blastn: 375 sarA_on_mibi1481.blastn: 248 * sarA_on_mibi1482.blastn: 375 sarA_on_mibi1483.blastn: 375 sarA_on_mibi1484.blastn: 375 sarA_on_mibi1485.blastn: 375 sarA_on_mibi1486.blastn: 375 sarA_on_mibi1487.blastn: 375 sarA_on_mibi1488.blastn: 375 sarA_on_mibi1489.blastn: 375 sarA_on_mibi1490.blastn: 375 sarA_on_mibi1491.blastn: 375 sarA_on_mibi1492.blastn: 375 sarA_on_mibi1493.blastn: 375 sarA_on_mibi1494.blastn: 375 sarA_on_mibi1495.blastn: 375 sarA_on_mibi1496.blastn: 375 sarA_on_mibi1497.blastn: 375 sarA_on_mibi1498.blastn: 375 sarA_on_mibi1499.blastn: 375 sarA_on_mibi1500.blastn: 375 sarA_on_mibi1501.blastn: 375 sarA_on_mibi1502.blastn: 375 sarA_on_mibi1503.blastn: 375 sarA_on_mibi1504.blastn: 375 sarA_on_mibi1505.blastn: 375 sarA_on_mibi1506.blastn: 357 sarA_on_mibi2312.blastn: 375 sarA_on_mibi2313.blastn: 375 sarA_on_mibi2314.blastn: 375 sarA_on_mibi2315.blastn: 375 sarA_on_mibi2316.blastn: 375 sarA_on_mibi2317.blastn: 375 sarA_on_mibi2318.blastn: 375 sarA_on_mibi2319.blastn: 375 sarA_on_mibi2320.blastn: 375 sarA_on_mibi2321.blastn: 375 sarA_on_mibi2379.blastn: 375 # -length(agrC.fasta)=1290 samtools faidx agrABCD_hld.fasta "gi|3320006|emb|Z49220.1|":1494-2783 for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query agrC.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > agrC_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py agrC_on_${sample}.blastn done agrC_on_mibi1435.blastn: 1002 agrC_on_mibi1436.blastn: 1290 agrC_on_mibi1437.blastn: 688 agrC_on_mibi1438.blastn: 1290 agrC_on_mibi1439.blastn: 688 agrC_on_mibi1440.blastn: 1290 agrC_on_mibi1441.blastn: 861 agrC_on_mibi1442.blastn: 688 agrC_on_mibi1443.blastn: 1290 agrC_on_mibi1444.blastn: 1290 agrC_on_mibi1445.blastn: 688 agrC_on_mibi1446.blastn: 688 agrC_on_mibi1447.blastn: 688 agrC_on_mibi1448.blastn: 1290 agrC_on_mibi1449.blastn: 861 agrC_on_mibi1450.blastn: 1290 agrC_on_mibi1451.blastn: 688 agrC_on_mibi1452.blastn: 818 agrC_on_mibi1453.blastn: 1290 agrC_on_mibi1454.blastn: 1290 agrC_on_mibi1455.blastn: 1290 agrC_on_mibi1456.blastn: 1290 agrC_on_mibi1457.blastn: 688 agrC_on_mibi1458.blastn: 1290 agrC_on_mibi1459.blastn: 1290 agrC_on_mibi1460.blastn: 688 agrC_on_mibi1461.blastn: 1290 agrC_on_mibi1462.blastn: 1290 agrC_on_mibi1463.blastn: 1290 agrC_on_mibi1464.blastn: 861 agrC_on_mibi1465.blastn: 688 agrC_on_mibi1466.blastn: 541 agrC_on_mibi1467.blastn: 688 agrC_on_mibi1468.blastn: 1290 agrC_on_mibi1469.blastn: 1290 agrC_on_mibi1471.blastn: 1165 agrC_on_mibi1473.blastn: 688 agrC_on_mibi1474.blastn: 1290 agrC_on_mibi1475.blastn: 688 agrC_on_mibi1476.blastn: 1290 agrC_on_mibi1477.blastn: 688 agrC_on_mibi1478.blastn: 442 agrC_on_mibi1479.blastn: 688 agrC_on_mibi1480.blastn: 1290 agrC_on_mibi1481.blastn: 862 agrC_on_mibi1482.blastn: 1290 agrC_on_mibi1483.blastn: 1290 agrC_on_mibi1484.blastn: 491 agrC_on_mibi1485.blastn: 1290 agrC_on_mibi1486.blastn: 1290 agrC_on_mibi1487.blastn: 1290 agrC_on_mibi1488.blastn: 1290 agrC_on_mibi1489.blastn: 688 agrC_on_mibi1490.blastn: 688 agrC_on_mibi1491.blastn: 688 agrC_on_mibi1492.blastn: 688 agrC_on_mibi1493.blastn: 818 agrC_on_mibi1494.blastn: 1290 agrC_on_mibi1495.blastn: 688 agrC_on_mibi1496.blastn: 1290 agrC_on_mibi1497.blastn: 911 agrC_on_mibi1498.blastn: 1290 agrC_on_mibi1499.blastn: 688 agrC_on_mibi1500.blastn: 1290 agrC_on_mibi1501.blastn: 1290 agrC_on_mibi1502.blastn: 1094 agrC_on_mibi1503.blastn: 818 agrC_on_mibi1504.blastn: 861 agrC_on_mibi1505.blastn: 1290 agrC_on_mibi1506.blastn: 1290 agrC_on_mibi2312.blastn: 688 agrC_on_mibi2313.blastn: 1290 agrC_on_mibi2314.blastn: 1290 agrC_on_mibi2315.blastn: 1290 agrC_on_mibi2316.blastn: 688 agrC_on_mibi2317.blastn: 688 agrC_on_mibi2318.blastn: 1290 agrC_on_mibi2319.blastn: 1290 agrC_on_mibi2320.blastn: 688 agrC_on_mibi2321.blastn: 1290 agrC_on_mibi2379.blastn: 688 partial, 1002/1290 nt present + partial, 688/1290 nt present + partial, 688/1290 nt present + partial, 861/1290 nt present partial, 688/1290 nt present + + partial, 688/1290 nt present partial, 688/1290 nt present partial, 688/1290 nt present + partial, 861/1290 nt present + partial, 688/1290 nt present partial, 818/1290 nt present + + + + partial, 688/1290 nt present + + partial, 688/1290 nt present + + + partial, 861/1290 nt present partial, 688/1290 nt present partial, 541/1290 nt present partial, 688/1290 nt present + + partial, 1165/1290 nt present partial, 688/1290 nt present + partial, 688/1290 nt present + partial, 688/1290 nt present partial, 442/1290 nt present partial, 688/1290 nt present + partial, 862/1290 nt present + + partial, 491/1290 nt present + + + + partial, 688/1290 nt present partial, 688/1290 nt present partial, 688/1290 nt present partial, 688/1290 nt present partial, 818/1290 nt present + partial, 688/1290 nt present + partial, 911/1290 nt present + partial, 688/1290 nt present + + partial, 1094/1290 nt present partial, 818/1290 nt present partial, 861/1290 nt present + + partial, 688/1290 nt present + + + partial, 688/1290 nt present partial, 688/1290 nt present + + partial, 688/1290 nt present + partial, 688/1290 nt present # - length(yycG.fasta)=1827 gene complement(2589240..2591072) /gene="yycG" samtools faidx CP000029.fasta "gi|57636010|gb|CP000029.1|":2589240-2591072 > yycG.fasta for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query yycG_revcomp.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > yycG_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py yycG_on_${sample}.blastn done #-->WARNING: Only 80% similarity, not the same gene! yycG_on_mibi1435.blastn: 1833 yycG_on_mibi1436.blastn: 1833 yycG_on_mibi1437.blastn: 1833 yycG_on_mibi1438.blastn: 1833 yycG_on_mibi1439.blastn: 1833 yycG_on_mibi1440.blastn: 1833 yycG_on_mibi1441.blastn: 1833 yycG_on_mibi1442.blastn: 1833 yycG_on_mibi1443.blastn: 1833 yycG_on_mibi1444.blastn: 1833 yycG_on_mibi1445.blastn: 1833 yycG_on_mibi1446.blastn: 1833 yycG_on_mibi1447.blastn: 1833 yycG_on_mibi1448.blastn: 1833 yycG_on_mibi1449.blastn: 1833 yycG_on_mibi1450.blastn: 1833 yycG_on_mibi1451.blastn: 1833 yycG_on_mibi1452.blastn: 1833 yycG_on_mibi1453.blastn: 1833 yycG_on_mibi1454.blastn: 1833 yycG_on_mibi1455.blastn: 1833 yycG_on_mibi1456.blastn: 1833 yycG_on_mibi1457.blastn: 1833 yycG_on_mibi1458.blastn: 1833 yycG_on_mibi1459.blastn: 1833 yycG_on_mibi1460.blastn: 1833 yycG_on_mibi1461.blastn: 1833 yycG_on_mibi1462.blastn: 1833 yycG_on_mibi1463.blastn: 1833 yycG_on_mibi1464.blastn: 1833 yycG_on_mibi1465.blastn: 1833 yycG_on_mibi1466.blastn: 1833 yycG_on_mibi1467.blastn: 1833 yycG_on_mibi1468.blastn: 1833 yycG_on_mibi1469.blastn: 1833 yycG_on_mibi1471.blastn: 1833 yycG_on_mibi1473.blastn: 1833 yycG_on_mibi1474.blastn: 1833 yycG_on_mibi1475.blastn: 1833 yycG_on_mibi1476.blastn: 1833 yycG_on_mibi1477.blastn: 1833 yycG_on_mibi1478.blastn: 1833 yycG_on_mibi1479.blastn: 1833 yycG_on_mibi1480.blastn: 1833 yycG_on_mibi1481.blastn: 1833 yycG_on_mibi1482.blastn: 1833 yycG_on_mibi1483.blastn: 1833 yycG_on_mibi1484.blastn: 1833 yycG_on_mibi1485.blastn: 1833 yycG_on_mibi1486.blastn: 1833 yycG_on_mibi1487.blastn: 1833 yycG_on_mibi1488.blastn: 1833 yycG_on_mibi1489.blastn: 1833 yycG_on_mibi1490.blastn: 1833 yycG_on_mibi1491.blastn: 1833 yycG_on_mibi1492.blastn: 1833 yycG_on_mibi1493.blastn: 1833 yycG_on_mibi1494.blastn: 1833 yycG_on_mibi1495.blastn: 1833 yycG_on_mibi1496.blastn: 1833 yycG_on_mibi1497.blastn: 1833 yycG_on_mibi1498.blastn: 1833 yycG_on_mibi1499.blastn: 1833 yycG_on_mibi1500.blastn: 1833 yycG_on_mibi1501.blastn: 1833 yycG_on_mibi1502.blastn: 1833 yycG_on_mibi1503.blastn: 1833 yycG_on_mibi1504.blastn: 1833 yycG_on_mibi1505.blastn: 1833 yycG_on_mibi1506.blastn: 1833 yycG_on_mibi2312.blastn: 1833 yycG_on_mibi2313.blastn: 1833 yycG_on_mibi2314.blastn: 1833 yycG_on_mibi2315.blastn: 1833 yycG_on_mibi2316.blastn: 1833 yycG_on_mibi2317.blastn: 1833 yycG_on_mibi2318.blastn: 1833 yycG_on_mibi2319.blastn: 1833 yycG_on_mibi2320.blastn: 1833 yycG_on_mibi2321.blastn: 1833 yycG_on_mibi2379.blastn: 1833 #TODOs: exact the gene sequence for psm-delta and atl #https://www.ncbi.nlm.nih.gov/nuccore (psm gene) AND "Staphylococcus aureus"[porgn:__txid1280] #grep "PSM-delta" mibi1435.gbff # /product="Phenol-soluble modulin PSM-delta" #grep "atl" mibi1435.gbff # /gene="atl" # /gene="atl" https://www.ncbi.nlm.nih.gov/nuccore/JQ066320.1 # -length(psm-beta1.fasta)=135 for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query psm-beta1.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > psm-beta1_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py psm-beta1_on_${sample}.blastn done psm-beta1_on_mibi1435.blastn: 0 psm-beta1_on_mibi1436.blastn: 0 psm-beta1_on_mibi1437.blastn: 0 psm-beta1_on_mibi1438.blastn: 0 psm-beta1_on_mibi1439.blastn: 0 psm-beta1_on_mibi1440.blastn: 0 psm-beta1_on_mibi1441.blastn: 0 psm-beta1_on_mibi1442.blastn: 0 psm-beta1_on_mibi1443.blastn: 0 psm-beta1_on_mibi1444.blastn: 0 psm-beta1_on_mibi1445.blastn: 0 psm-beta1_on_mibi1446.blastn: 0 psm-beta1_on_mibi1447.blastn: 0 psm-beta1_on_mibi1448.blastn: 0 psm-beta1_on_mibi1449.blastn: 0 psm-beta1_on_mibi1450.blastn: 0 psm-beta1_on_mibi1451.blastn: 0 psm-beta1_on_mibi1452.blastn: 0 psm-beta1_on_mibi1453.blastn: 0 psm-beta1_on_mibi1454.blastn: 0 psm-beta1_on_mibi1455.blastn: 0 psm-beta1_on_mibi1456.blastn: 0 psm-beta1_on_mibi1457.blastn: 0 psm-beta1_on_mibi1458.blastn: 0 psm-beta1_on_mibi1459.blastn: 0 psm-beta1_on_mibi1460.blastn: 0 psm-beta1_on_mibi1461.blastn: 0 psm-beta1_on_mibi1462.blastn: 0 psm-beta1_on_mibi1463.blastn: 0 psm-beta1_on_mibi1464.blastn: 0 psm-beta1_on_mibi1465.blastn: 0 psm-beta1_on_mibi1466.blastn: 0 psm-beta1_on_mibi1467.blastn: 0 psm-beta1_on_mibi1468.blastn: 0 psm-beta1_on_mibi1469.blastn: 0 psm-beta1_on_mibi1471.blastn: 0 psm-beta1_on_mibi1473.blastn: 0 psm-beta1_on_mibi1474.blastn: 0 psm-beta1_on_mibi1475.blastn: 0 psm-beta1_on_mibi1476.blastn: 0 psm-beta1_on_mibi1477.blastn: 0 psm-beta1_on_mibi1478.blastn: 0 psm-beta1_on_mibi1479.blastn: 0 psm-beta1_on_mibi1480.blastn: 0 psm-beta1_on_mibi1481.blastn: 0 psm-beta1_on_mibi1482.blastn: 0 psm-beta1_on_mibi1483.blastn: 0 psm-beta1_on_mibi1484.blastn: 0 psm-beta1_on_mibi1485.blastn: 0 psm-beta1_on_mibi1486.blastn: 0 psm-beta1_on_mibi1487.blastn: 0 psm-beta1_on_mibi1488.blastn: 0 psm-beta1_on_mibi1489.blastn: 0 psm-beta1_on_mibi1490.blastn: 0 psm-beta1_on_mibi1491.blastn: 0 psm-beta1_on_mibi1492.blastn: 0 psm-beta1_on_mibi1493.blastn: 0 psm-beta1_on_mibi1494.blastn: 0 psm-beta1_on_mibi1495.blastn: 0 psm-beta1_on_mibi1496.blastn: 0 psm-beta1_on_mibi1497.blastn: 0 psm-beta1_on_mibi1498.blastn: 0 psm-beta1_on_mibi1499.blastn: 0 psm-beta1_on_mibi1500.blastn: 0 psm-beta1_on_mibi1501.blastn: 0 psm-beta1_on_mibi1502.blastn: 0 psm-beta1_on_mibi1503.blastn: 0 psm-beta1_on_mibi1504.blastn: 0 psm-beta1_on_mibi1505.blastn: 0 psm-beta1_on_mibi1506.blastn: 0 psm-beta1_on_mibi2312.blastn: 0 psm-beta1_on_mibi2313.blastn: 0 psm-beta1_on_mibi2314.blastn: 0 psm-beta1_on_mibi2315.blastn: 0 psm-beta1_on_mibi2316.blastn: 0 psm-beta1_on_mibi2317.blastn: 0 psm-beta1_on_mibi2318.blastn: 0 psm-beta1_on_mibi2319.blastn: 0 psm-beta1_on_mibi2320.blastn: 0 psm-beta1_on_mibi2321.blastn: 0 psm-beta1_on_mibi2379.blastn: 0 # -length(hlb.fasta)=993 for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query psm-beta.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > hlb_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py hlb_on_${sample}.blastn done hlb_on_mibi1435.blastn: 0 hlb_on_mibi1436.blastn: 0 hlb_on_mibi1437.blastn: 0 hlb_on_mibi1438.blastn: 0 hlb_on_mibi1439.blastn: 0 hlb_on_mibi1440.blastn: 0 hlb_on_mibi1441.blastn: 0 hlb_on_mibi1442.blastn: 0 hlb_on_mibi1443.blastn: 0 hlb_on_mibi1444.blastn: 0 hlb_on_mibi1445.blastn: 0 hlb_on_mibi1446.blastn: 0 hlb_on_mibi1447.blastn: 0 hlb_on_mibi1448.blastn: 0 hlb_on_mibi1449.blastn: 0 hlb_on_mibi1450.blastn: 0 hlb_on_mibi1451.blastn: 0 hlb_on_mibi1452.blastn: 0 hlb_on_mibi1453.blastn: 0 hlb_on_mibi1454.blastn: 0 hlb_on_mibi1455.blastn: 0 hlb_on_mibi1456.blastn: 0 hlb_on_mibi1457.blastn: 0 hlb_on_mibi1458.blastn: 0 hlb_on_mibi1459.blastn: 0 hlb_on_mibi1460.blastn: 0 hlb_on_mibi1461.blastn: 0 hlb_on_mibi1462.blastn: 0 hlb_on_mibi1463.blastn: 0 hlb_on_mibi1464.blastn: 0 hlb_on_mibi1465.blastn: 0 hlb_on_mibi1466.blastn: 0 hlb_on_mibi1467.blastn: 0 hlb_on_mibi1468.blastn: 0 hlb_on_mibi1469.blastn: 0 hlb_on_mibi1471.blastn: 0 hlb_on_mibi1473.blastn: 0 hlb_on_mibi1474.blastn: 0 hlb_on_mibi1475.blastn: 0 hlb_on_mibi1476.blastn: 0 hlb_on_mibi1477.blastn: 0 hlb_on_mibi1478.blastn: 0 hlb_on_mibi1479.blastn: 0 hlb_on_mibi1480.blastn: 0 hlb_on_mibi1481.blastn: 0 hlb_on_mibi1482.blastn: 0 hlb_on_mibi1483.blastn: 0 hlb_on_mibi1484.blastn: 0 hlb_on_mibi1485.blastn: 0 hlb_on_mibi1486.blastn: 0 hlb_on_mibi1487.blastn: 0 hlb_on_mibi1488.blastn: 0 hlb_on_mibi1489.blastn: 0 hlb_on_mibi1490.blastn: 0 hlb_on_mibi1491.blastn: 0 hlb_on_mibi1492.blastn: 0 hlb_on_mibi1493.blastn: 0 hlb_on_mibi1494.blastn: 0 hlb_on_mibi1495.blastn: 0 hlb_on_mibi1496.blastn: 0 hlb_on_mibi1497.blastn: 0 hlb_on_mibi1498.blastn: 0 hlb_on_mibi1499.blastn: 0 hlb_on_mibi1500.blastn: 0 hlb_on_mibi1501.blastn: 0 hlb_on_mibi1502.blastn: 0 hlb_on_mibi1503.blastn: 0 hlb_on_mibi1504.blastn: 0 hlb_on_mibi1505.blastn: 0 hlb_on_mibi1506.blastn: 0 hlb_on_mibi2312.blastn: 0 hlb_on_mibi2313.blastn: 0 hlb_on_mibi2314.blastn: 0 hlb_on_mibi2315.blastn: 0 hlb_on_mibi2316.blastn: 0 hlb_on_mibi2317.blastn: 0 hlb_on_mibi2318.blastn: 0 hlb_on_mibi2319.blastn: 0 hlb_on_mibi2320.blastn: 0 hlb_on_mibi2321.blastn: 0 hlb_on_mibi2379.blastn: 0 ## -length(altE_revcomp.fasta)=4008 #gene complement(627656..631663) # /gene="atlE" #samtools faidx CP000029.fasta gi|57636010|gb|CP000029.1| samtools faidx CP000029.fasta "gi|57636010|gb|CP000029.1|":627656-631663 > atlE.fasta revcomp atlE.fasta > atlE_revcomp.fasta for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query atlE_revcomp.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > atlE_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py atlE_on_${sample}.blastn done atlE_on_mibi1435.blastn: 4008 atlE_on_mibi1436.blastn: 4008 atlE_on_mibi1437.blastn: 4465 atlE_on_mibi1438.blastn: 4008 atlE_on_mibi1439.blastn: 4008 atlE_on_mibi1440.blastn: 4008 atlE_on_mibi1441.blastn: 4450 atlE_on_mibi1442.blastn: 4450 atlE_on_mibi1443.blastn: 4008 atlE_on_mibi1444.blastn: 4008 atlE_on_mibi1445.blastn: 4008 atlE_on_mibi1446.blastn: 4008 atlE_on_mibi1447.blastn: 4008 atlE_on_mibi1448.blastn: 4008 atlE_on_mibi1449.blastn: 4008 atlE_on_mibi1450.blastn: 4008 atlE_on_mibi1451.blastn: 4008 atlE_on_mibi1452.blastn: 4450 atlE_on_mibi1453.blastn: 4008 atlE_on_mibi1454.blastn: 4008 atlE_on_mibi1455.blastn: 4008 atlE_on_mibi1456.blastn: 4008 atlE_on_mibi1457.blastn: 4008 atlE_on_mibi1458.blastn: 4450 atlE_on_mibi1459.blastn: 4008 atlE_on_mibi1460.blastn: 4008 atlE_on_mibi1461.blastn: 4008 atlE_on_mibi1462.blastn: 4008 atlE_on_mibi1463.blastn: 4450 atlE_on_mibi1464.blastn: 4450 atlE_on_mibi1465.blastn: 4008 atlE_on_mibi1466.blastn: 4008 atlE_on_mibi1467.blastn: 4450 atlE_on_mibi1468.blastn: 4465 atlE_on_mibi1469.blastn: 4465 atlE_on_mibi1471.blastn: 4008 atlE_on_mibi1473.blastn: 4008 atlE_on_mibi1474.blastn: 4008 atlE_on_mibi1475.blastn: 4008 atlE_on_mibi1476.blastn: 4008 atlE_on_mibi1477.blastn: 4008 atlE_on_mibi1478.blastn: 4008 atlE_on_mibi1479.blastn: 4008 atlE_on_mibi1480.blastn: 4008 atlE_on_mibi1481.blastn: 4008 atlE_on_mibi1482.blastn: 4008 atlE_on_mibi1483.blastn: 4450 atlE_on_mibi1484.blastn: 4008 atlE_on_mibi1485.blastn: 4008 atlE_on_mibi1486.blastn: 4450 atlE_on_mibi1487.blastn: 4008 atlE_on_mibi1488.blastn: 4008 atlE_on_mibi1489.blastn: 4450 atlE_on_mibi1490.blastn: 4008 atlE_on_mibi1491.blastn: 4008 atlE_on_mibi1492.blastn: 4008 atlE_on_mibi1493.blastn: 4450 atlE_on_mibi1494.blastn: 4008 atlE_on_mibi1495.blastn: 4008 atlE_on_mibi1496.blastn: 4008 atlE_on_mibi1497.blastn: 4008 atlE_on_mibi1498.blastn: 4008 atlE_on_mibi1499.blastn: 4008 atlE_on_mibi1500.blastn: 4008 atlE_on_mibi1501.blastn: 4008 atlE_on_mibi1502.blastn: 4008 atlE_on_mibi1503.blastn: 4008 atlE_on_mibi1504.blastn: 4450 atlE_on_mibi1505.blastn: 4008 atlE_on_mibi1506.blastn: 4008 atlE_on_mibi2312.blastn: 4008 atlE_on_mibi2313.blastn: 4008 atlE_on_mibi2314.blastn: 4008 atlE_on_mibi2315.blastn: 4008 atlE_on_mibi2316.blastn: 4008 atlE_on_mibi2317.blastn: 4008 atlE_on_mibi2318.blastn: 4008 atlE_on_mibi2319.blastn: 4008 atlE_on_mibi2320.blastn: 4008 atlE_on_mibi2321.blastn: 4008 atlE_on_mibi2379.blastn: 4008 # -length(sdrG)=2796 for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query sdrG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sdrG_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py sdrG_on_${sample}.blastn done sdrG_on_mibi1435.blastn: 2973 sdrG_on_mibi1436.blastn: 2967 sdrG_on_mibi1437.blastn: 2821 sdrG_on_mibi1438.blastn: 2796 sdrG_on_mibi1439.blastn: 3155 sdrG_on_mibi1440.blastn: 2967 sdrG_on_mibi1441.blastn: 2797 sdrG_on_mibi1442.blastn: 2801 sdrG_on_mibi1443.blastn: 2966 sdrG_on_mibi1444.blastn: 2796 sdrG_on_mibi1445.blastn: 3158 sdrG_on_mibi1446.blastn: 3155 sdrG_on_mibi1447.blastn: 2966 sdrG_on_mibi1448.blastn: 2797 sdrG_on_mibi1449.blastn: 2712 sdrG_on_mibi1450.blastn: 2794 sdrG_on_mibi1451.blastn: 2946 sdrG_on_mibi1452.blastn: 3166 sdrG_on_mibi1453.blastn: 2970 sdrG_on_mibi1454.blastn: 2976 sdrG_on_mibi1455.blastn: 2797 sdrG_on_mibi1456.blastn: 2967 sdrG_on_mibi1457.blastn: 2963 sdrG_on_mibi1458.blastn: 2794 sdrG_on_mibi1459.blastn: 3153 sdrG_on_mibi1460.blastn: 2566 sdrG_on_mibi1461.blastn: 2500 sdrG_on_mibi1462.blastn: 2966 sdrG_on_mibi1463.blastn: 2971 sdrG_on_mibi1464.blastn: 2530 sdrG_on_mibi1465.blastn: 2981 sdrG_on_mibi1466.blastn: 1547 * sdrG_on_mibi1467.blastn: 2794 sdrG_on_mibi1468.blastn: 2967 sdrG_on_mibi1469.blastn: 2967 sdrG_on_mibi1471.blastn: 2796 sdrG_on_mibi1473.blastn: 2976 sdrG_on_mibi1474.blastn: 2797 sdrG_on_mibi1475.blastn: 2981 sdrG_on_mibi1476.blastn: 2794 sdrG_on_mibi1477.blastn: 2967 sdrG_on_mibi1478.blastn: 1511 * sdrG_on_mibi1479.blastn: 2966 sdrG_on_mibi1480.blastn: 2976 sdrG_on_mibi1481.blastn: 2797 sdrG_on_mibi1482.blastn: 2797 sdrG_on_mibi1483.blastn: 2797 sdrG_on_mibi1484.blastn: 2976 sdrG_on_mibi1485.blastn: 3001 sdrG_on_mibi1486.blastn: 2797 sdrG_on_mibi1487.blastn: 2976 sdrG_on_mibi1488.blastn: 2963 sdrG_on_mibi1489.blastn: 2797 sdrG_on_mibi1490.blastn: 2981 sdrG_on_mibi1491.blastn: 3155 sdrG_on_mibi1492.blastn: 2982 sdrG_on_mibi1493.blastn: 2904 sdrG_on_mibi1494.blastn: 2980 sdrG_on_mibi1495.blastn: 2976 sdrG_on_mibi1496.blastn: 2994 sdrG_on_mibi1497.blastn: 2797 sdrG_on_mibi1498.blastn: 2967 sdrG_on_mibi1499.blastn: 2976 sdrG_on_mibi1500.blastn: 2797 sdrG_on_mibi1501.blastn: 2797 sdrG_on_mibi1502.blastn: 2976 sdrG_on_mibi1503.blastn: 2976 sdrG_on_mibi1504.blastn: 2634 sdrG_on_mibi1505.blastn: 2970 sdrG_on_mibi1506.blastn: 2985 sdrG_on_mibi2312.blastn: 2866 sdrG_on_mibi2313.blastn: 2794 sdrG_on_mibi2314.blastn: 3171 sdrG_on_mibi2315.blastn: 2797 sdrG_on_mibi2316.blastn: 3158 sdrG_on_mibi2317.blastn: 2975 sdrG_on_mibi2318.blastn: 2976 sdrG_on_mibi2319.blastn: 2613 sdrG_on_mibi2320.blastn: 2972 sdrG_on_mibi2321.blastn: 2794 sdrG_on_mibi2379.blastn: 2975 ##length(sdrH)=1446 for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query sdrH_revcomp.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sdrH_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py sdrH_on_${sample}.blastn done sdrH_on_mibi1435.blastn: 1647 sdrH_on_mibi1436.blastn: 1683 sdrH_on_mibi1437.blastn: 1482 sdrH_on_mibi1438.blastn: 1608 sdrH_on_mibi1439.blastn: 1739 sdrH_on_mibi1440.blastn: 1644 sdrH_on_mibi1441.blastn: 1446 sdrH_on_mibi1442.blastn: 1482 sdrH_on_mibi1443.blastn: 1683 sdrH_on_mibi1444.blastn: 1683 sdrH_on_mibi1445.blastn: 1482 sdrH_on_mibi1446.blastn: 1482 sdrH_on_mibi1447.blastn: 1482 sdrH_on_mibi1448.blastn: 1446 sdrH_on_mibi1449.blastn: 1450 sdrH_on_mibi1450.blastn: 1446 sdrH_on_mibi1451.blastn: 1487 sdrH_on_mibi1452.blastn: 1446 sdrH_on_mibi1453.blastn: 1464 sdrH_on_mibi1454.blastn: 1647 sdrH_on_mibi1455.blastn: 1644 sdrH_on_mibi1456.blastn: 1647 sdrH_on_mibi1457.blastn: 1572 sdrH_on_mibi1458.blastn: 1632 sdrH_on_mibi1459.blastn: 1647 sdrH_on_mibi1460.blastn: 1482 sdrH_on_mibi1461.blastn: 1647 sdrH_on_mibi1462.blastn: 1683 sdrH_on_mibi1463.blastn: 1647 sdrH_on_mibi1464.blastn: 1739 sdrH_on_mibi1465.blastn: 1464 sdrH_on_mibi1466.blastn: 1305 sdrH_on_mibi1467.blastn: 1482 sdrH_on_mibi1468.blastn: 1559 sdrH_on_mibi1469.blastn: 1556 sdrH_on_mibi1471.blastn: 1572 sdrH_on_mibi1473.blastn: 1482 sdrH_on_mibi1474.blastn: 1446 sdrH_on_mibi1475.blastn: 1482 sdrH_on_mibi1476.blastn: 1458 sdrH_on_mibi1477.blastn: 1647 sdrH_on_mibi1478.blastn: 710 * sdrH_on_mibi1479.blastn: 1482 sdrH_on_mibi1480.blastn: 1647 sdrH_on_mibi1481.blastn: 1412 sdrH_on_mibi1482.blastn: 1647 sdrH_on_mibi1483.blastn: 1647 sdrH_on_mibi1484.blastn: 1448 sdrH_on_mibi1485.blastn: 1647 sdrH_on_mibi1486.blastn: 1827 sdrH_on_mibi1487.blastn: 1647 sdrH_on_mibi1488.blastn: 1683 sdrH_on_mibi1489.blastn: 1739 sdrH_on_mibi1490.blastn: 1482 sdrH_on_mibi1491.blastn: 1482 sdrH_on_mibi1492.blastn: 1482 sdrH_on_mibi1493.blastn: 1440 sdrH_on_mibi1494.blastn: 1458 sdrH_on_mibi1495.blastn: 1482 sdrH_on_mibi1496.blastn: 1644 sdrH_on_mibi1497.blastn: 1736 sdrH_on_mibi1498.blastn: 1647 sdrH_on_mibi1499.blastn: 1482 sdrH_on_mibi1500.blastn: 1683 sdrH_on_mibi1501.blastn: 1647 sdrH_on_mibi1502.blastn: 1647 sdrH_on_mibi1503.blastn: 1745 sdrH_on_mibi1504.blastn: 1683 sdrH_on_mibi1505.blastn: 1736 sdrH_on_mibi1506.blastn: 1683 sdrH_on_mibi2312.blastn: 1556 sdrH_on_mibi2313.blastn: 1464 sdrH_on_mibi2314.blastn: 1464 sdrH_on_mibi2315.blastn: 1683 sdrH_on_mibi2316.blastn: 1482 sdrH_on_mibi2317.blastn: 1464 sdrH_on_mibi2318.blastn: 1647 sdrH_on_mibi2319.blastn: 1644 sdrH_on_mibi2320.blastn: 1464 sdrH_on_mibi2321.blastn: 1464 sdrH_on_mibi2379.blastn: 1647 # -length(ebh_revcomp.fasta)=30450 for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query ebh_revcomp.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ebh_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py ebh_on_${sample}.blastn done ebh_on_mibi1435.blastn: 30450 ebh_on_mibi1436.blastn: 30450 ebh_on_mibi1437.blastn: 29490 ebh_on_mibi1438.blastn: 25428 ebh_on_mibi1439.blastn: 30452 ebh_on_mibi1440.blastn: 11933 ebh_on_mibi1441.blastn: 30450 ebh_on_mibi1442.blastn: 30463 ebh_on_mibi1443.blastn: 30450 ebh_on_mibi1444.blastn: 30450 ebh_on_mibi1445.blastn: 30450 ebh_on_mibi1446.blastn: 30450 ebh_on_mibi1447.blastn: 30450 ebh_on_mibi1448.blastn: 30450 ebh_on_mibi1449.blastn: 30450 ebh_on_mibi1450.blastn: 30450 ebh_on_mibi1451.blastn: 30450 ebh_on_mibi1452.blastn: 21401 ebh_on_mibi1453.blastn: 30450 ebh_on_mibi1454.blastn: 11933 ebh_on_mibi1455.blastn: 30450 ebh_on_mibi1456.blastn: 11933 ebh_on_mibi1457.blastn: 30450 ebh_on_mibi1458.blastn: 29955 ebh_on_mibi1459.blastn: 11933 ebh_on_mibi1460.blastn: 30450 ebh_on_mibi1461.blastn: 11933 ebh_on_mibi1462.blastn: 30450 ebh_on_mibi1463.blastn: 20602 ebh_on_mibi1464.blastn: 30450 ebh_on_mibi1465.blastn: 30450 ebh_on_mibi1466.blastn: 14964 ebh_on_mibi1467.blastn: 30463 ebh_on_mibi1468.blastn: 30450 ebh_on_mibi1469.blastn: 30450 ebh_on_mibi1471.blastn: 25428 ebh_on_mibi1473.blastn: 30450 ebh_on_mibi1474.blastn: 30450 ebh_on_mibi1475.blastn: 30450 ebh_on_mibi1476.blastn: 30450 ebh_on_mibi1477.blastn: 30384 ebh_on_mibi1478.blastn: 11933 ebh_on_mibi1479.blastn: 30450 ebh_on_mibi1480.blastn: 11933 ebh_on_mibi1481.blastn: 25694 ebh_on_mibi1482.blastn: 30450 ebh_on_mibi1483.blastn: 30450 ebh_on_mibi1484.blastn: 30450 ebh_on_mibi1485.blastn: 11933 ebh_on_mibi1486.blastn: 30450 ebh_on_mibi1487.blastn: 11933 ebh_on_mibi1488.blastn: 30450 ebh_on_mibi1489.blastn: 30450 ebh_on_mibi1490.blastn: 30450 ebh_on_mibi1491.blastn: 30450 ebh_on_mibi1492.blastn: 30450 ebh_on_mibi1493.blastn: 30465 ebh_on_mibi1494.blastn: 30450 ebh_on_mibi1495.blastn: 30450 ebh_on_mibi1496.blastn: 11933 ebh_on_mibi1497.blastn: 30450 ebh_on_mibi1498.blastn: 11933 ebh_on_mibi1499.blastn: 30450 ebh_on_mibi1500.blastn: 30450 ebh_on_mibi1501.blastn: 30450 ebh_on_mibi1502.blastn: 11933 ebh_on_mibi1503.blastn: 30450 ebh_on_mibi1504.blastn: 30450 ebh_on_mibi1505.blastn: 30450 ebh_on_mibi1506.blastn: 30450 ebh_on_mibi2312.blastn: 30450 ebh_on_mibi2313.blastn: 30450 ebh_on_mibi2314.blastn: 30450 ebh_on_mibi2315.blastn: 30461 ebh_on_mibi2316.blastn: 30450 ebh_on_mibi2317.blastn: 30450 ebh_on_mibi2318.blastn: 11933 ebh_on_mibi2319.blastn: 11933 ebh_on_mibi2320.blastn: 30110 ebh_on_mibi2321.blastn: 30450 ebh_on_mibi2379.blastn: 30450 #For gene: ebpS, tagB, capC, sepA, dltA, fmtC # -length(SE0760)=1383 for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query ebpS_revcomp.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ebpS_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py ebpS_on_${sample}.blastn done ebpS_on_mibi1435.blastn: 1383 ebpS_on_mibi1436.blastn: 1383 ebpS_on_mibi1437.blastn: 1383 ebpS_on_mibi1438.blastn: 1383 ebpS_on_mibi1439.blastn: 1383 ebpS_on_mibi1440.blastn: 1383 ebpS_on_mibi1441.blastn: 1383 ebpS_on_mibi1442.blastn: 1408 ebpS_on_mibi1443.blastn: 1383 ebpS_on_mibi1444.blastn: 1383 ebpS_on_mibi1445.blastn: 1383 ebpS_on_mibi1446.blastn: 1383 ebpS_on_mibi1447.blastn: 1383 ebpS_on_mibi1448.blastn: 1383 ebpS_on_mibi1449.blastn: 1383 ebpS_on_mibi1450.blastn: 1383 ebpS_on_mibi1451.blastn: 1383 ebpS_on_mibi1452.blastn: 1408 ebpS_on_mibi1453.blastn: 1383 ebpS_on_mibi1454.blastn: 1383 ebpS_on_mibi1455.blastn: 1383 ebpS_on_mibi1456.blastn: 1383 ebpS_on_mibi1457.blastn: 1383 ebpS_on_mibi1458.blastn: 1408 ebpS_on_mibi1459.blastn: 1383 ebpS_on_mibi1460.blastn: 1383 ebpS_on_mibi1461.blastn: 1383 ebpS_on_mibi1462.blastn: 1383 ebpS_on_mibi1463.blastn: 1383 ebpS_on_mibi1464.blastn: 1392 ebpS_on_mibi1465.blastn: 1383 ebpS_on_mibi1466.blastn: 1383 ebpS_on_mibi1467.blastn: 1420 ebpS_on_mibi1468.blastn: 1383 ebpS_on_mibi1469.blastn: 1383 ebpS_on_mibi1471.blastn: 1383 ebpS_on_mibi1473.blastn: 1383 ebpS_on_mibi1474.blastn: 1383 ebpS_on_mibi1475.blastn: 1383 ebpS_on_mibi1476.blastn: 1383 ebpS_on_mibi1477.blastn: 1275 ebpS_on_mibi1478.blastn: 1383 ebpS_on_mibi1479.blastn: 1383 ebpS_on_mibi1480.blastn: 1383 ebpS_on_mibi1481.blastn: 1383 ebpS_on_mibi1482.blastn: 1383 ebpS_on_mibi1483.blastn: 1383 ebpS_on_mibi1484.blastn: 1383 ebpS_on_mibi1485.blastn: 1383 ebpS_on_mibi1486.blastn: 1383 ebpS_on_mibi1487.blastn: 1383 ebpS_on_mibi1488.blastn: 1383 ebpS_on_mibi1489.blastn: 1383 ebpS_on_mibi1490.blastn: 1383 ebpS_on_mibi1491.blastn: 1383 ebpS_on_mibi1492.blastn: 1383 ebpS_on_mibi1493.blastn: 1420 ebpS_on_mibi1494.blastn: 1383 ebpS_on_mibi1495.blastn: 1383 ebpS_on_mibi1496.blastn: 1383 ebpS_on_mibi1497.blastn: 1383 ebpS_on_mibi1498.blastn: 1383 ebpS_on_mibi1499.blastn: 1383 ebpS_on_mibi1500.blastn: 1383 ebpS_on_mibi1501.blastn: 1383 ebpS_on_mibi1502.blastn: 1383 ebpS_on_mibi1503.blastn: 1383 ebpS_on_mibi1504.blastn: 1408 ebpS_on_mibi1505.blastn: 1383 ebpS_on_mibi1506.blastn: 1383 ebpS_on_mibi2312.blastn: 1383 ebpS_on_mibi2313.blastn: 1383 ebpS_on_mibi2314.blastn: 1383 ebpS_on_mibi2315.blastn: 1383 ebpS_on_mibi2316.blastn: 1383 ebpS_on_mibi2317.blastn: 1383 ebpS_on_mibi2318.blastn: 1383 ebpS_on_mibi2319.blastn: 1383 ebpS_on_mibi2320.blastn: 1383 ebpS_on_mibi2321.blastn: 1383 ebpS_on_mibi2379.blastn: 1383 # -length(tagB)=1680 #mibi1435.gff3:contig_2 Prodigal CDS 146190 147869 . - 0 ID=MMJCKL_03035;Name=CDP-glycerol glycerophosphotransferase%2C TagB/SpsB family;locus_tag=MMJCKL_03035;product=CDP-glycerol glycerophosphotransferase%2C TagB/SpsB family;Dbxref=COG:COG1887,COG:MI,RefSeq:WP_001831509.1,SO:0001217,UniParc:UPI00004BA7A8,UniRef:UniRef100_A0A4Q9W688,UniRef:UniRef50_A0A0H2VI21,UniRef:UniRef90_A0A0H2VI21;gene=tagB samtools faidx mibi1435.fna contig_2:146190-147869 > tagB_mibi1435.fasta revcomp tagB_mibi1435.fasta > tagB_mibi1435_revcomp.fasta for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query tagB_mibi1435_revcomp.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > tagB_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py tagB_on_${sample}.blastn done tagB_on_mibi1435.blastn: 1680 tagB_on_mibi1436.blastn: 1680 tagB_on_mibi1437.blastn: 1680 tagB_on_mibi1438.blastn: 1680 tagB_on_mibi1439.blastn: 1680 tagB_on_mibi1440.blastn: 1680 tagB_on_mibi1441.blastn: 1680 tagB_on_mibi1442.blastn: 1680 tagB_on_mibi1443.blastn: 1680 tagB_on_mibi1444.blastn: 1680 tagB_on_mibi1445.blastn: 1680 tagB_on_mibi1446.blastn: 1680 tagB_on_mibi1447.blastn: 1680 tagB_on_mibi1448.blastn: 1680 tagB_on_mibi1449.blastn: 1680 tagB_on_mibi1450.blastn: 1680 tagB_on_mibi1451.blastn: 1680 tagB_on_mibi1452.blastn: 1680 tagB_on_mibi1453.blastn: 1680 tagB_on_mibi1454.blastn: 1680 tagB_on_mibi1455.blastn: 1680 tagB_on_mibi1456.blastn: 1680 tagB_on_mibi1457.blastn: 1680 tagB_on_mibi1458.blastn: 1680 tagB_on_mibi1459.blastn: 1680 tagB_on_mibi1460.blastn: 1680 tagB_on_mibi1461.blastn: 1680 tagB_on_mibi1462.blastn: 1680 tagB_on_mibi1463.blastn: 1680 tagB_on_mibi1464.blastn: 1680 tagB_on_mibi1465.blastn: 1680 tagB_on_mibi1466.blastn: 1680 tagB_on_mibi1467.blastn: 1680 tagB_on_mibi1468.blastn: 1680 tagB_on_mibi1469.blastn: 1680 tagB_on_mibi1471.blastn: 1680 tagB_on_mibi1473.blastn: 1680 tagB_on_mibi1474.blastn: 1680 tagB_on_mibi1475.blastn: 1680 tagB_on_mibi1476.blastn: 1680 tagB_on_mibi1477.blastn: 1680 tagB_on_mibi1478.blastn: 1680 tagB_on_mibi1479.blastn: 1680 tagB_on_mibi1480.blastn: 1680 tagB_on_mibi1481.blastn: 1680 tagB_on_mibi1482.blastn: 1680 tagB_on_mibi1483.blastn: 1680 tagB_on_mibi1484.blastn: 1680 tagB_on_mibi1485.blastn: 1680 tagB_on_mibi1486.blastn: 1680 tagB_on_mibi1487.blastn: 1680 tagB_on_mibi1488.blastn: 1680 tagB_on_mibi1489.blastn: 1680 tagB_on_mibi1490.blastn: 1680 tagB_on_mibi1491.blastn: 1680 tagB_on_mibi1492.blastn: 1680 tagB_on_mibi1493.blastn: 1680 tagB_on_mibi1494.blastn: 1680 tagB_on_mibi1495.blastn: 1680 tagB_on_mibi1496.blastn: 1680 tagB_on_mibi1497.blastn: 1680 tagB_on_mibi1498.blastn: 1680 tagB_on_mibi1499.blastn: 1680 tagB_on_mibi1500.blastn: 1680 tagB_on_mibi1501.blastn: 1680 tagB_on_mibi1502.blastn: 1680 tagB_on_mibi1503.blastn: 1680 tagB_on_mibi1504.blastn: 1680 tagB_on_mibi1505.blastn: 1680 tagB_on_mibi1506.blastn: 1680 tagB_on_mibi2312.blastn: 1680 tagB_on_mibi2313.blastn: 1680 tagB_on_mibi2314.blastn: 1680 tagB_on_mibi2315.blastn: 1680 tagB_on_mibi2316.blastn: 1680 tagB_on_mibi2317.blastn: 1680 tagB_on_mibi2318.blastn: 1680 tagB_on_mibi2319.blastn: 1680 tagB_on_mibi2320.blastn: 1680 tagB_on_mibi2321.blastn: 1680 tagB_on_mibi2379.blastn: 1680 # -length(capC)=450 for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query capC.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > capC_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py capC_on_${sample}.blastn done capC_on_mibi1435.blastn: 0 capC_on_mibi1436.blastn: 0 capC_on_mibi1437.blastn: 0 capC_on_mibi1438.blastn: 0 capC_on_mibi1439.blastn: 0 capC_on_mibi1440.blastn: 0 capC_on_mibi1441.blastn: 0 capC_on_mibi1442.blastn: 0 capC_on_mibi1443.blastn: 0 capC_on_mibi1444.blastn: 0 capC_on_mibi1445.blastn: 0 capC_on_mibi1446.blastn: 0 capC_on_mibi1447.blastn: 0 capC_on_mibi1448.blastn: 0 capC_on_mibi1449.blastn: 0 capC_on_mibi1450.blastn: 0 capC_on_mibi1451.blastn: 0 capC_on_mibi1452.blastn: 0 capC_on_mibi1453.blastn: 0 capC_on_mibi1454.blastn: 0 capC_on_mibi1455.blastn: 0 capC_on_mibi1456.blastn: 0 capC_on_mibi1457.blastn: 0 capC_on_mibi1458.blastn: 0 capC_on_mibi1459.blastn: 0 capC_on_mibi1460.blastn: 0 capC_on_mibi1461.blastn: 0 capC_on_mibi1462.blastn: 0 capC_on_mibi1463.blastn: 0 capC_on_mibi1464.blastn: 0 capC_on_mibi1465.blastn: 0 capC_on_mibi1466.blastn: 0 capC_on_mibi1467.blastn: 0 capC_on_mibi1468.blastn: 0 capC_on_mibi1469.blastn: 0 capC_on_mibi1471.blastn: 0 capC_on_mibi1473.blastn: 0 capC_on_mibi1474.blastn: 0 capC_on_mibi1475.blastn: 0 capC_on_mibi1476.blastn: 0 capC_on_mibi1477.blastn: 0 capC_on_mibi1478.blastn: 0 capC_on_mibi1479.blastn: 0 capC_on_mibi1480.blastn: 0 capC_on_mibi1481.blastn: 0 capC_on_mibi1482.blastn: 0 capC_on_mibi1483.blastn: 0 capC_on_mibi1484.blastn: 0 capC_on_mibi1485.blastn: 0 capC_on_mibi1486.blastn: 0 capC_on_mibi1487.blastn: 0 capC_on_mibi1488.blastn: 0 capC_on_mibi1489.blastn: 0 capC_on_mibi1490.blastn: 0 capC_on_mibi1491.blastn: 0 capC_on_mibi1492.blastn: 0 capC_on_mibi1493.blastn: 0 capC_on_mibi1494.blastn: 0 capC_on_mibi1495.blastn: 0 capC_on_mibi1496.blastn: 0 capC_on_mibi1497.blastn: 0 capC_on_mibi1498.blastn: 0 capC_on_mibi1499.blastn: 0 capC_on_mibi1500.blastn: 0 capC_on_mibi1501.blastn: 0 capC_on_mibi1502.blastn: 0 capC_on_mibi1503.blastn: 0 capC_on_mibi1504.blastn: 0 capC_on_mibi1505.blastn: 0 capC_on_mibi1506.blastn: 0 capC_on_mibi2312.blastn: 0 capC_on_mibi2313.blastn: 0 capC_on_mibi2314.blastn: 0 capC_on_mibi2315.blastn: 0 capC_on_mibi2316.blastn: 0 capC_on_mibi2317.blastn: 0 capC_on_mibi2318.blastn: 0 capC_on_mibi2319.blastn: 0 capC_on_mibi2320.blastn: 0 capC_on_mibi2321.blastn: 0 capC_on_mibi2379.blastn: 0 # -length(sepA_mibi1435.fasta)=1524 #contig_7 Prodigal CDS 52151 53674 . + 0 ID=MMJCKL_06215;Name=Extracellular elastase;locus_tag=MMJCKL_06215;product=Extracellular elastase;Dbxref=COG:COG3227,COG:O,EC:3.4.24.-,GO:0004222,GO:0005576,GO:0006508,GO:0046872,KEGG:K01401,RefSeq:WP_002486087.1,SO:0001217,UniParc:UPI00024E17FE,UniRef:UniRef100_UPI00066C9785,UniRef:UniRef50_P0C0Q3,UniRef:UniRef90_P0C0Q3;gene=sepA samtools faidx mibi1435.fna contig_7:52151-53674 > sepA_mibi1435.fasta for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query sepA_mibi1435.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sepA_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py sepA_on_${sample}.blastn done sepA_on_mibi1435.blastn: 1524 sepA_on_mibi1436.blastn: 1524 sepA_on_mibi1437.blastn: 1523 sepA_on_mibi1438.blastn: 1524 sepA_on_mibi1439.blastn: 1523 sepA_on_mibi1440.blastn: 1523 sepA_on_mibi1441.blastn: 1523 sepA_on_mibi1442.blastn: 1523 sepA_on_mibi1443.blastn: 1523 sepA_on_mibi1444.blastn: 1523 sepA_on_mibi1445.blastn: 1523 sepA_on_mibi1446.blastn: 1523 sepA_on_mibi1447.blastn: 1523 sepA_on_mibi1448.blastn: 1524 sepA_on_mibi1449.blastn: 1523 sepA_on_mibi1450.blastn: 1523 sepA_on_mibi1451.blastn: 1523 sepA_on_mibi1452.blastn: 1523 sepA_on_mibi1453.blastn: 1523 sepA_on_mibi1454.blastn: 1523 sepA_on_mibi1455.blastn: 1524 sepA_on_mibi1456.blastn: 1523 sepA_on_mibi1457.blastn: 1523 sepA_on_mibi1458.blastn: 1523 sepA_on_mibi1459.blastn: 1523 sepA_on_mibi1460.blastn: 1523 sepA_on_mibi1461.blastn: 1523 sepA_on_mibi1462.blastn: 1523 sepA_on_mibi1463.blastn: 1523 sepA_on_mibi1464.blastn: 1523 sepA_on_mibi1465.blastn: 1523 sepA_on_mibi1466.blastn: 1523 sepA_on_mibi1467.blastn: 1524 sepA_on_mibi1468.blastn: 1523 sepA_on_mibi1469.blastn: 1523 sepA_on_mibi1471.blastn: 1524 sepA_on_mibi1473.blastn: 1523 sepA_on_mibi1474.blastn: 1524 sepA_on_mibi1475.blastn: 1523 sepA_on_mibi1476.blastn: 1523 sepA_on_mibi1477.blastn: 1524 sepA_on_mibi1478.blastn: 1523 sepA_on_mibi1479.blastn: 1523 sepA_on_mibi1480.blastn: 1523 sepA_on_mibi1481.blastn: 1524 sepA_on_mibi1482.blastn: 1524 sepA_on_mibi1483.blastn: 1523 sepA_on_mibi1484.blastn: 1523 sepA_on_mibi1485.blastn: 1523 sepA_on_mibi1486.blastn: 1523 sepA_on_mibi1487.blastn: 1523 sepA_on_mibi1488.blastn: 1523 sepA_on_mibi1489.blastn: 1523 sepA_on_mibi1490.blastn: 1523 sepA_on_mibi1491.blastn: 1523 sepA_on_mibi1492.blastn: 1523 sepA_on_mibi1493.blastn: 1523 sepA_on_mibi1494.blastn: 1523 sepA_on_mibi1495.blastn: 1523 sepA_on_mibi1496.blastn: 1523 sepA_on_mibi1497.blastn: 1523 sepA_on_mibi1498.blastn: 1523 sepA_on_mibi1499.blastn: 1523 sepA_on_mibi1500.blastn: 1523 sepA_on_mibi1501.blastn: 1523 sepA_on_mibi1502.blastn: 1523 sepA_on_mibi1503.blastn: 1523 sepA_on_mibi1504.blastn: 1523 sepA_on_mibi1505.blastn: 1523 sepA_on_mibi1506.blastn: 1523 sepA_on_mibi2312.blastn: 1523 sepA_on_mibi2313.blastn: 1523 sepA_on_mibi2314.blastn: 1523 sepA_on_mibi2315.blastn: 1523 sepA_on_mibi2316.blastn: 1523 sepA_on_mibi2317.blastn: 1523 sepA_on_mibi2318.blastn: 1523 sepA_on_mibi2319.blastn: 1523 sepA_on_mibi2320.blastn: 1523 sepA_on_mibi2321.blastn: 1523 sepA_on_mibi2379.blastn: 1523 # -length(dltA.fasta)=1458 for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query dltA.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > dltA_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py dltA_on_${sample}.blastn done dltA_on_mibi1435.blastn: 1458 dltA_on_mibi1436.blastn: 1458 dltA_on_mibi1437.blastn: 1458 dltA_on_mibi1438.blastn: 1458 dltA_on_mibi1439.blastn: 1458 dltA_on_mibi1440.blastn: 1458 dltA_on_mibi1441.blastn: 1458 dltA_on_mibi1442.blastn: 1458 dltA_on_mibi1443.blastn: 1458 dltA_on_mibi1444.blastn: 1458 dltA_on_mibi1445.blastn: 1458 dltA_on_mibi1446.blastn: 1458 dltA_on_mibi1447.blastn: 1458 dltA_on_mibi1448.blastn: 1464 dltA_on_mibi1449.blastn: 1458 dltA_on_mibi1450.blastn: 1458 dltA_on_mibi1451.blastn: 1458 dltA_on_mibi1452.blastn: 1458 dltA_on_mibi1453.blastn: 1458 dltA_on_mibi1454.blastn: 1458 dltA_on_mibi1455.blastn: 1458 dltA_on_mibi1456.blastn: 1458 dltA_on_mibi1457.blastn: 1458 dltA_on_mibi1458.blastn: 1458 dltA_on_mibi1459.blastn: 1458 dltA_on_mibi1460.blastn: 1458 dltA_on_mibi1461.blastn: 1458 dltA_on_mibi1462.blastn: 1458 dltA_on_mibi1463.blastn: 1458 dltA_on_mibi1464.blastn: 1458 dltA_on_mibi1465.blastn: 1458 dltA_on_mibi1466.blastn: 1458 dltA_on_mibi1467.blastn: 1458 dltA_on_mibi1468.blastn: 1458 dltA_on_mibi1469.blastn: 1458 dltA_on_mibi1471.blastn: 1458 dltA_on_mibi1473.blastn: 1458 dltA_on_mibi1474.blastn: 1458 dltA_on_mibi1475.blastn: 1458 dltA_on_mibi1476.blastn: 1458 dltA_on_mibi1477.blastn: 1458 dltA_on_mibi1478.blastn: 1458 dltA_on_mibi1479.blastn: 1458 dltA_on_mibi1480.blastn: 1458 dltA_on_mibi1481.blastn: 1458 dltA_on_mibi1482.blastn: 1458 dltA_on_mibi1483.blastn: 1458 dltA_on_mibi1484.blastn: 1458 dltA_on_mibi1485.blastn: 1458 dltA_on_mibi1486.blastn: 1458 dltA_on_mibi1487.blastn: 1458 dltA_on_mibi1488.blastn: 1458 dltA_on_mibi1489.blastn: 1458 dltA_on_mibi1490.blastn: 1458 dltA_on_mibi1491.blastn: 1458 dltA_on_mibi1492.blastn: 1458 dltA_on_mibi1493.blastn: 1458 dltA_on_mibi1494.blastn: 1458 dltA_on_mibi1495.blastn: 1458 dltA_on_mibi1496.blastn: 1458 dltA_on_mibi1497.blastn: 1458 dltA_on_mibi1498.blastn: 1458 dltA_on_mibi1499.blastn: 1458 dltA_on_mibi1500.blastn: 1458 dltA_on_mibi1501.blastn: 1458 dltA_on_mibi1502.blastn: 1458 dltA_on_mibi1503.blastn: 1458 dltA_on_mibi1504.blastn: 1458 dltA_on_mibi1505.blastn: 1458 dltA_on_mibi1506.blastn: 1458 dltA_on_mibi2312.blastn: 1458 dltA_on_mibi2313.blastn: 1458 dltA_on_mibi2314.blastn: 1458 dltA_on_mibi2315.blastn: 1458 dltA_on_mibi2316.blastn: 1458 dltA_on_mibi2317.blastn: 1458 dltA_on_mibi2318.blastn: 1458 dltA_on_mibi2319.blastn: 1458 dltA_on_mibi2320.blastn: 1458 dltA_on_mibi2321.blastn: 1458 dltA_on_mibi2379.blastn: 1458 # -length(fmtC.fasta)=2523 for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query fmtC.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > fmtC_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py fmtC_on_${sample}.blastn done fmtC_on_mibi1435.blastn: 0 fmtC_on_mibi1436.blastn: 0 fmtC_on_mibi1437.blastn: 0 fmtC_on_mibi1438.blastn: 0 fmtC_on_mibi1439.blastn: 0 fmtC_on_mibi1440.blastn: 0 fmtC_on_mibi1441.blastn: 0 fmtC_on_mibi1442.blastn: 0 fmtC_on_mibi1443.blastn: 0 fmtC_on_mibi1444.blastn: 0 fmtC_on_mibi1445.blastn: 0 fmtC_on_mibi1446.blastn: 0 fmtC_on_mibi1447.blastn: 0 fmtC_on_mibi1448.blastn: 0 fmtC_on_mibi1449.blastn: 0 fmtC_on_mibi1450.blastn: 0 fmtC_on_mibi1451.blastn: 0 fmtC_on_mibi1452.blastn: 0 fmtC_on_mibi1453.blastn: 0 fmtC_on_mibi1454.blastn: 0 fmtC_on_mibi1455.blastn: 0 fmtC_on_mibi1456.blastn: 0 fmtC_on_mibi1457.blastn: 0 fmtC_on_mibi1458.blastn: 0 fmtC_on_mibi1459.blastn: 0 fmtC_on_mibi1460.blastn: 0 fmtC_on_mibi1461.blastn: 0 fmtC_on_mibi1462.blastn: 0 fmtC_on_mibi1463.blastn: 0 fmtC_on_mibi1464.blastn: 0 fmtC_on_mibi1465.blastn: 0 fmtC_on_mibi1466.blastn: 0 fmtC_on_mibi1467.blastn: 0 fmtC_on_mibi1468.blastn: 0 fmtC_on_mibi1469.blastn: 0 fmtC_on_mibi1471.blastn: 0 fmtC_on_mibi1473.blastn: 0 fmtC_on_mibi1474.blastn: 0 fmtC_on_mibi1475.blastn: 0 fmtC_on_mibi1476.blastn: 0 fmtC_on_mibi1477.blastn: 0 fmtC_on_mibi1478.blastn: 0 fmtC_on_mibi1479.blastn: 0 fmtC_on_mibi1480.blastn: 0 fmtC_on_mibi1481.blastn: 0 fmtC_on_mibi1482.blastn: 0 fmtC_on_mibi1483.blastn: 0 fmtC_on_mibi1484.blastn: 0 fmtC_on_mibi1485.blastn: 0 fmtC_on_mibi1486.blastn: 0 fmtC_on_mibi1487.blastn: 0 fmtC_on_mibi1488.blastn: 0 fmtC_on_mibi1489.blastn: 0 fmtC_on_mibi1490.blastn: 0 fmtC_on_mibi1491.blastn: 0 fmtC_on_mibi1492.blastn: 0 fmtC_on_mibi1493.blastn: 0 fmtC_on_mibi1494.blastn: 0 fmtC_on_mibi1495.blastn: 0 fmtC_on_mibi1496.blastn: 0 fmtC_on_mibi1497.blastn: 0 fmtC_on_mibi1498.blastn: 0 fmtC_on_mibi1499.blastn: 0 fmtC_on_mibi1500.blastn: 0 fmtC_on_mibi1501.blastn: 0 fmtC_on_mibi1502.blastn: 0 fmtC_on_mibi1503.blastn: 0 fmtC_on_mibi1504.blastn: 0 fmtC_on_mibi1505.blastn: 0 fmtC_on_mibi1506.blastn: 0 fmtC_on_mibi2312.blastn: 0 fmtC_on_mibi2313.blastn: 0 fmtC_on_mibi2314.blastn: 0 fmtC_on_mibi2315.blastn: 0 fmtC_on_mibi2316.blastn: 0 fmtC_on_mibi2317.blastn: 0 fmtC_on_mibi2318.blastn: 0 fmtC_on_mibi2319.blastn: 0 fmtC_on_mibi2320.blastn: 0 fmtC_on_mibi2321.blastn: 0 fmtC_on_mibi2379.blastn: 0 #-- For genes: lipA (915), sceD (699), esp (660), ecpA (1188) for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query lipA.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./lipA_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py lipA_on_${sample}.blastn done lipA_on_mibi1435.blastn: 915 lipA_on_mibi1436.blastn: 915 lipA_on_mibi1437.blastn: 915 lipA_on_mibi1438.blastn: 915 lipA_on_mibi1439.blastn: 915 lipA_on_mibi1440.blastn: 915 lipA_on_mibi1441.blastn: 915 lipA_on_mibi1442.blastn: 915 lipA_on_mibi1443.blastn: 915 lipA_on_mibi1444.blastn: 915 lipA_on_mibi1445.blastn: 915 lipA_on_mibi1446.blastn: 915 lipA_on_mibi1447.blastn: 915 lipA_on_mibi1448.blastn: 915 lipA_on_mibi1449.blastn: 915 lipA_on_mibi1450.blastn: 915 lipA_on_mibi1451.blastn: 915 lipA_on_mibi1452.blastn: 915 lipA_on_mibi1453.blastn: 915 lipA_on_mibi1454.blastn: 915 lipA_on_mibi1455.blastn: 915 lipA_on_mibi1456.blastn: 915 lipA_on_mibi1457.blastn: 915 lipA_on_mibi1458.blastn: 915 lipA_on_mibi1459.blastn: 915 lipA_on_mibi1460.blastn: 915 lipA_on_mibi1461.blastn: 915 lipA_on_mibi1462.blastn: 915 lipA_on_mibi1463.blastn: 915 lipA_on_mibi1464.blastn: 915 lipA_on_mibi1465.blastn: 915 lipA_on_mibi1466.blastn: 915 lipA_on_mibi1467.blastn: 915 lipA_on_mibi1468.blastn: 915 lipA_on_mibi1469.blastn: 915 lipA_on_mibi1471.blastn: 915 lipA_on_mibi1473.blastn: 915 lipA_on_mibi1474.blastn: 915 lipA_on_mibi1475.blastn: 915 lipA_on_mibi1476.blastn: 915 lipA_on_mibi1477.blastn: 915 lipA_on_mibi1478.blastn: 915 lipA_on_mibi1479.blastn: 915 lipA_on_mibi1480.blastn: 915 lipA_on_mibi1481.blastn: 915 lipA_on_mibi1482.blastn: 915 lipA_on_mibi1483.blastn: 915 lipA_on_mibi1484.blastn: 915 lipA_on_mibi1485.blastn: 915 lipA_on_mibi1486.blastn: 915 lipA_on_mibi1487.blastn: 915 lipA_on_mibi1488.blastn: 915 lipA_on_mibi1489.blastn: 915 lipA_on_mibi1490.blastn: 915 lipA_on_mibi1491.blastn: 915 lipA_on_mibi1492.blastn: 915 lipA_on_mibi1493.blastn: 915 lipA_on_mibi1494.blastn: 915 lipA_on_mibi1495.blastn: 915 lipA_on_mibi1496.blastn: 915 lipA_on_mibi1497.blastn: 915 lipA_on_mibi1498.blastn: 915 lipA_on_mibi1499.blastn: 915 lipA_on_mibi1500.blastn: 915 lipA_on_mibi1501.blastn: 915 lipA_on_mibi1502.blastn: 915 lipA_on_mibi1503.blastn: 915 lipA_on_mibi1504.blastn: 915 lipA_on_mibi1505.blastn: 915 lipA_on_mibi1506.blastn: 915 lipA_on_mibi2312.blastn: 915 lipA_on_mibi2313.blastn: 915 lipA_on_mibi2314.blastn: 915 lipA_on_mibi2315.blastn: 915 lipA_on_mibi2316.blastn: 915 lipA_on_mibi2317.blastn: 915 lipA_on_mibi2318.blastn: 915 lipA_on_mibi2319.blastn: 915 lipA_on_mibi2320.blastn: 915 lipA_on_mibi2321.blastn: 915 lipA_on_mibi2379.blastn: 915 #grep "gene=sceD" mibi1435.gff3 #contig_11 Prodigal CDS 25537 26196 . - 0 ID=MMJCKL_08095;Name=putative transglycosylase SceD;locus_tag=MMJCKL_08095;product=putative transglycosylase SceD;Dbxref=EC:3.2.-.-,GO:0005576,GO:0008152,GO:0016798,RefSeq:WP_001829904.1,SO:0001217,UniParc:UPI000003B952,UniRef:UniRef100_Q5HMC6,UniRef:UniRef50_Q5HEA4,UniRef:UniRef90_Q5HMC6;gene=sceD samtools faidx mibi1435.fna contig_11:25537-26196 > sceD_mibi1435.fasta revcomp sceD_mibi1435.fasta > sceD_mibi1435_revcomp.fasta cp sceD_mibi1435_revcomp.fasta ../presence_absence_matrix_on_gene_list/ for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query sceD_mibi1435_revcomp.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./sceD_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py sceD_on_${sample}.blastn done sceD_on_mibi1435.blastn: 660 sceD_on_mibi1436.blastn: 660 sceD_on_mibi1437.blastn: 660 sceD_on_mibi1438.blastn: 660 sceD_on_mibi1439.blastn: 660 sceD_on_mibi1440.blastn: 660 sceD_on_mibi1441.blastn: 660 sceD_on_mibi1442.blastn: 660 sceD_on_mibi1443.blastn: 660 sceD_on_mibi1444.blastn: 660 sceD_on_mibi1445.blastn: 660 sceD_on_mibi1446.blastn: 660 sceD_on_mibi1447.blastn: 660 sceD_on_mibi1448.blastn: 660 sceD_on_mibi1449.blastn: 660 sceD_on_mibi1450.blastn: 660 sceD_on_mibi1451.blastn: 660 sceD_on_mibi1452.blastn: 660 sceD_on_mibi1453.blastn: 660 sceD_on_mibi1454.blastn: 660 sceD_on_mibi1455.blastn: 660 sceD_on_mibi1456.blastn: 660 sceD_on_mibi1457.blastn: 660 sceD_on_mibi1458.blastn: 660 sceD_on_mibi1459.blastn: 660 sceD_on_mibi1460.blastn: 660 sceD_on_mibi1461.blastn: 660 sceD_on_mibi1462.blastn: 660 sceD_on_mibi1463.blastn: 660 sceD_on_mibi1464.blastn: 660 sceD_on_mibi1465.blastn: 660 sceD_on_mibi1466.blastn: 660 sceD_on_mibi1467.blastn: 660 sceD_on_mibi1468.blastn: 660 sceD_on_mibi1469.blastn: 660 sceD_on_mibi1471.blastn: 660 sceD_on_mibi1473.blastn: 660 sceD_on_mibi1474.blastn: 660 sceD_on_mibi1475.blastn: 660 sceD_on_mibi1476.blastn: 660 sceD_on_mibi1477.blastn: 660 sceD_on_mibi1478.blastn: 660 sceD_on_mibi1479.blastn: 660 sceD_on_mibi1480.blastn: 660 sceD_on_mibi1481.blastn: 660 sceD_on_mibi1482.blastn: 660 sceD_on_mibi1483.blastn: 660 sceD_on_mibi1484.blastn: 660 sceD_on_mibi1485.blastn: 660 sceD_on_mibi1486.blastn: 660 sceD_on_mibi1487.blastn: 660 sceD_on_mibi1488.blastn: 660 sceD_on_mibi1489.blastn: 660 sceD_on_mibi1490.blastn: 660 sceD_on_mibi1491.blastn: 660 sceD_on_mibi1492.blastn: 660 sceD_on_mibi1493.blastn: 660 sceD_on_mibi1494.blastn: 660 sceD_on_mibi1495.blastn: 660 sceD_on_mibi1496.blastn: 660 sceD_on_mibi1497.blastn: 660 sceD_on_mibi1498.blastn: 660 sceD_on_mibi1499.blastn: 660 sceD_on_mibi1500.blastn: 660 sceD_on_mibi1501.blastn: 660 sceD_on_mibi1502.blastn: 660 sceD_on_mibi1503.blastn: 660 sceD_on_mibi1504.blastn: 660 sceD_on_mibi1505.blastn: 660 sceD_on_mibi1506.blastn: 660 sceD_on_mibi2312.blastn: 660 sceD_on_mibi2313.blastn: 660 sceD_on_mibi2314.blastn: 660 sceD_on_mibi2315.blastn: 660 sceD_on_mibi2316.blastn: 660 sceD_on_mibi2317.blastn: 660 sceD_on_mibi2318.blastn: 660 sceD_on_mibi2319.blastn: 660 sceD_on_mibi2320.blastn: 660 sceD_on_mibi2321.blastn: 660 sceD_on_mibi2379.blastn: 660 for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query esp.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./esp_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py esp_on_${sample}.blastn done esp_on_mibi1435.blastn: 0 esp_on_mibi1436.blastn: 0 esp_on_mibi1437.blastn: 0 esp_on_mibi1438.blastn: 0 esp_on_mibi1439.blastn: 0 esp_on_mibi1440.blastn: 0 esp_on_mibi1441.blastn: 0 esp_on_mibi1442.blastn: 0 esp_on_mibi1443.blastn: 0 esp_on_mibi1444.blastn: 0 esp_on_mibi1445.blastn: 0 esp_on_mibi1446.blastn: 0 esp_on_mibi1447.blastn: 0 esp_on_mibi1448.blastn: 0 esp_on_mibi1449.blastn: 0 esp_on_mibi1450.blastn: 0 esp_on_mibi1451.blastn: 0 esp_on_mibi1452.blastn: 0 esp_on_mibi1453.blastn: 0 esp_on_mibi1454.blastn: 0 esp_on_mibi1455.blastn: 0 esp_on_mibi1456.blastn: 0 esp_on_mibi1457.blastn: 0 esp_on_mibi1458.blastn: 0 esp_on_mibi1459.blastn: 0 esp_on_mibi1460.blastn: 0 esp_on_mibi1461.blastn: 0 esp_on_mibi1462.blastn: 0 esp_on_mibi1463.blastn: 0 esp_on_mibi1464.blastn: 0 esp_on_mibi1465.blastn: 0 esp_on_mibi1466.blastn: 0 esp_on_mibi1467.blastn: 0 esp_on_mibi1468.blastn: 0 esp_on_mibi1469.blastn: 0 esp_on_mibi1471.blastn: 0 esp_on_mibi1473.blastn: 0 esp_on_mibi1474.blastn: 0 esp_on_mibi1475.blastn: 0 esp_on_mibi1476.blastn: 0 esp_on_mibi1477.blastn: 0 esp_on_mibi1478.blastn: 0 esp_on_mibi1479.blastn: 0 esp_on_mibi1480.blastn: 0 esp_on_mibi1481.blastn: 0 esp_on_mibi1482.blastn: 0 esp_on_mibi1483.blastn: 0 esp_on_mibi1484.blastn: 0 esp_on_mibi1485.blastn: 0 esp_on_mibi1486.blastn: 0 esp_on_mibi1487.blastn: 0 esp_on_mibi1488.blastn: 0 esp_on_mibi1489.blastn: 0 esp_on_mibi1490.blastn: 0 esp_on_mibi1491.blastn: 0 esp_on_mibi1492.blastn: 0 esp_on_mibi1493.blastn: 0 esp_on_mibi1494.blastn: 0 esp_on_mibi1495.blastn: 0 esp_on_mibi1496.blastn: 0 esp_on_mibi1497.blastn: 0 esp_on_mibi1498.blastn: 0 esp_on_mibi1499.blastn: 0 esp_on_mibi1500.blastn: 0 esp_on_mibi1501.blastn: 0 esp_on_mibi1502.blastn: 0 esp_on_mibi1503.blastn: 0 esp_on_mibi1504.blastn: 0 esp_on_mibi1505.blastn: 0 esp_on_mibi1506.blastn: 0 esp_on_mibi2312.blastn: 0 esp_on_mibi2313.blastn: 0 esp_on_mibi2314.blastn: 0 esp_on_mibi2315.blastn: 0 esp_on_mibi2316.blastn: 0 esp_on_mibi2317.blastn: 0 esp_on_mibi2318.blastn: 0 esp_on_mibi2319.blastn: 0 esp_on_mibi2320.blastn: 0 esp_on_mibi2321.blastn: 0 esp_on_mibi2379.blastn: 0 #grep "gene=ecpA" mibi1435.gff3 #contig_9 Prodigal CDS 6382 7569 . - 0 ID=MMJCKL_07070;Name=Extracellular cysteine protease;locus_tag=MMJCKL_07070;product=Extracellular cysteine protease;Dbxref=EC:3.4.22.-,GO:0005576,GO:0006508,GO:0008234,KEGG:K08258,RefSeq:WP_002497714.1,SO:0001217,UniParc:UPI00026C1A6F,UniRef:UniRef100_UPI00026C1A6F,UniRef:UniRef50_P81297,UniRef:UniRef90_Q5HKF6;gene=ecpA samtools faidx mibi1435.fna contig_9:6382-7569 > ecpA_mibi1435.fasta revcomp ecpA_mibi1435.fasta > ecpA_mibi1435_revcomp.fasta cp ecpA_mibi1435_revcomp.fasta ../presence_absence_matrix_on_gene_list/ for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query ecpA_mibi1435_revcomp.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./ecpA_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py ecpA_on_${sample}.blastn done ecpA_on_mibi1435.blastn: 1188 ecpA_on_mibi1436.blastn: 1188 ecpA_on_mibi1437.blastn: 1188 ecpA_on_mibi1438.blastn: 1188 ecpA_on_mibi1439.blastn: 1188 ecpA_on_mibi1440.blastn: 1188 ecpA_on_mibi1441.blastn: 1188 ecpA_on_mibi1442.blastn: 1188 ecpA_on_mibi1443.blastn: 1188 ecpA_on_mibi1444.blastn: 1188 ecpA_on_mibi1445.blastn: 1188 ecpA_on_mibi1446.blastn: 1188 ecpA_on_mibi1447.blastn: 1188 ecpA_on_mibi1448.blastn: 1188 ecpA_on_mibi1449.blastn: 1188 ecpA_on_mibi1450.blastn: 1188 ecpA_on_mibi1451.blastn: 1188 ecpA_on_mibi1452.blastn: 1188 ecpA_on_mibi1453.blastn: 1188 ecpA_on_mibi1454.blastn: 1188 ecpA_on_mibi1455.blastn: 1188 ecpA_on_mibi1456.blastn: 1188 ecpA_on_mibi1457.blastn: 1188 ecpA_on_mibi1458.blastn: 1188 ecpA_on_mibi1459.blastn: 1188 ecpA_on_mibi1460.blastn: 1188 ecpA_on_mibi1461.blastn: 1188 ecpA_on_mibi1462.blastn: 1188 ecpA_on_mibi1463.blastn: 1188 ecpA_on_mibi1464.blastn: 1188 ecpA_on_mibi1465.blastn: 1188 ecpA_on_mibi1466.blastn: 1188 ecpA_on_mibi1467.blastn: 1188 ecpA_on_mibi1468.blastn: 1188 ecpA_on_mibi1469.blastn: 1188 ecpA_on_mibi1471.blastn: 1188 ecpA_on_mibi1473.blastn: 1188 ecpA_on_mibi1474.blastn: 1188 ecpA_on_mibi1475.blastn: 1188 ecpA_on_mibi1476.blastn: 1188 ecpA_on_mibi1477.blastn: 1188 ecpA_on_mibi1478.blastn: 1188 ecpA_on_mibi1479.blastn: 1188 ecpA_on_mibi1480.blastn: 1188 ecpA_on_mibi1481.blastn: 1188 ecpA_on_mibi1482.blastn: 1188 ecpA_on_mibi1483.blastn: 1188 ecpA_on_mibi1484.blastn: 1188 ecpA_on_mibi1485.blastn: 1188 ecpA_on_mibi1486.blastn: 1188 ecpA_on_mibi1487.blastn: 1188 ecpA_on_mibi1488.blastn: 1188 ecpA_on_mibi1489.blastn: 1188 ecpA_on_mibi1490.blastn: 1188 ecpA_on_mibi1491.blastn: 1188 ecpA_on_mibi1492.blastn: 1188 ecpA_on_mibi1493.blastn: 1188 ecpA_on_mibi1494.blastn: 1188 ecpA_on_mibi1495.blastn: 1188 ecpA_on_mibi1496.blastn: 1188 ecpA_on_mibi1497.blastn: 1188 ecpA_on_mibi1498.blastn: 1188 ecpA_on_mibi1499.blastn: 1188 ecpA_on_mibi1500.blastn: 1188 ecpA_on_mibi1501.blastn: 1188 ecpA_on_mibi1502.blastn: 1188 ecpA_on_mibi1503.blastn: 1188 ecpA_on_mibi1504.blastn: 1188 ecpA_on_mibi1505.blastn: 1188 ecpA_on_mibi1506.blastn: 1188 ecpA_on_mibi2312.blastn: 1188 ecpA_on_mibi2313.blastn: 1188 ecpA_on_mibi2314.blastn: 1188 ecpA_on_mibi2315.blastn: 1188 ecpA_on_mibi2316.blastn: 1188 ecpA_on_mibi2317.blastn: 1188 ecpA_on_mibi2318.blastn: 1188 ecpA_on_mibi2319.blastn: 1188 ecpA_on_mibi2320.blastn: 1188 ecpA_on_mibi2321.blastn: 1188 ecpA_on_mibi2379.blastn: 1188 # -length(SE0760)=798 for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query SE0760.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > SE0760_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py SE0760_on_${sample}.blastn done SE0760_on_mibi1435.blastn: 798 SE0760_on_mibi1436.blastn: 798 SE0760_on_mibi1437.blastn: 798 SE0760_on_mibi1438.blastn: 798 SE0760_on_mibi1439.blastn: 798 SE0760_on_mibi1440.blastn: 798 SE0760_on_mibi1441.blastn: 798 SE0760_on_mibi1442.blastn: 798 SE0760_on_mibi1443.blastn: 798 SE0760_on_mibi1444.blastn: 798 SE0760_on_mibi1445.blastn: 798 SE0760_on_mibi1446.blastn: 798 SE0760_on_mibi1447.blastn: 798 SE0760_on_mibi1448.blastn: 798 SE0760_on_mibi1449.blastn: 798 SE0760_on_mibi1450.blastn: 798 SE0760_on_mibi1451.blastn: 798 SE0760_on_mibi1452.blastn: 798 SE0760_on_mibi1453.blastn: 798 SE0760_on_mibi1454.blastn: 798 SE0760_on_mibi1455.blastn: 798 SE0760_on_mibi1456.blastn: 798 SE0760_on_mibi1457.blastn: 798 SE0760_on_mibi1458.blastn: 798 SE0760_on_mibi1459.blastn: 798 SE0760_on_mibi1460.blastn: 798 SE0760_on_mibi1461.blastn: 798 SE0760_on_mibi1462.blastn: 798 SE0760_on_mibi1463.blastn: 798 SE0760_on_mibi1464.blastn: 798 SE0760_on_mibi1465.blastn: 798 SE0760_on_mibi1466.blastn: 798 SE0760_on_mibi1467.blastn: 799 SE0760_on_mibi1468.blastn: 798 SE0760_on_mibi1469.blastn: 798 SE0760_on_mibi1471.blastn: 798 SE0760_on_mibi1473.blastn: 798 SE0760_on_mibi1474.blastn: 798 SE0760_on_mibi1475.blastn: 798 SE0760_on_mibi1476.blastn: 798 SE0760_on_mibi1477.blastn: 798 SE0760_on_mibi1478.blastn: 798 SE0760_on_mibi1479.blastn: 798 SE0760_on_mibi1480.blastn: 798 SE0760_on_mibi1481.blastn: 798 SE0760_on_mibi1482.blastn: 798 SE0760_on_mibi1483.blastn: 798 SE0760_on_mibi1484.blastn: 798 SE0760_on_mibi1485.blastn: 798 SE0760_on_mibi1486.blastn: 798 SE0760_on_mibi1487.blastn: 798 SE0760_on_mibi1488.blastn: 798 SE0760_on_mibi1489.blastn: 798 SE0760_on_mibi1490.blastn: 798 SE0760_on_mibi1491.blastn: 798 SE0760_on_mibi1492.blastn: 798 SE0760_on_mibi1493.blastn: 798 SE0760_on_mibi1494.blastn: 798 SE0760_on_mibi1495.blastn: 798 SE0760_on_mibi1496.blastn: 798 SE0760_on_mibi1497.blastn: 798 SE0760_on_mibi1498.blastn: 798 SE0760_on_mibi1499.blastn: 798 SE0760_on_mibi1500.blastn: 798 SE0760_on_mibi1501.blastn: 798 SE0760_on_mibi1502.blastn: 798 SE0760_on_mibi1503.blastn: 798 SE0760_on_mibi1504.blastn: 798 SE0760_on_mibi1505.blastn: 798 SE0760_on_mibi1506.blastn: 798 SE0760_on_mibi2312.blastn: 798 SE0760_on_mibi2313.blastn: 798 SE0760_on_mibi2314.blastn: 798 SE0760_on_mibi2315.blastn: 798 SE0760_on_mibi2316.blastn: 798 SE0760_on_mibi2317.blastn: 798 SE0760_on_mibi2318.blastn: 798 SE0760_on_mibi2319.blastn: 798 SE0760_on_mibi2320.blastn: 798 SE0760_on_mibi2321.blastn: 798 SE0760_on_mibi2379.blastn: 798 # -lenth(MT880870)=34053 for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query MT880870.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./MT880870_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py MT880870_on_${sample}.blastn done MT880870_on_mibi1435.blastn: 2366 MT880870_on_mibi1436.blastn: 3198 MT880870_on_mibi1437.blastn: 3198 MT880870_on_mibi1438.blastn: 2366 MT880870_on_mibi1439.blastn: 3198 MT880870_on_mibi1440.blastn: 34177 MT880870_on_mibi1441.blastn: 2372 MT880870_on_mibi1442.blastn: 3470 MT880870_on_mibi1443.blastn: 2366 MT880870_on_mibi1444.blastn: 2372 MT880870_on_mibi1445.blastn: 3198 MT880870_on_mibi1446.blastn: 3198 MT880870_on_mibi1447.blastn: 3198 MT880870_on_mibi1448.blastn: 3198 MT880870_on_mibi1449.blastn: 2366 MT880870_on_mibi1450.blastn: 3198 MT880870_on_mibi1451.blastn: 1085 MT880870_on_mibi1452.blastn: 1832 MT880870_on_mibi1453.blastn: 2366 MT880870_on_mibi1454.blastn: 34177 MT880870_on_mibi1455.blastn: 3198 MT880870_on_mibi1456.blastn: 34177 MT880870_on_mibi1457.blastn: 3198 MT880870_on_mibi1458.blastn: 835 MT880870_on_mibi1459.blastn: 34177 MT880870_on_mibi1460.blastn: 3198 MT880870_on_mibi1461.blastn: 34177 MT880870_on_mibi1462.blastn: 2372 MT880870_on_mibi1463.blastn: 2372 MT880870_on_mibi1464.blastn: 2311 MT880870_on_mibi1465.blastn: 3198 MT880870_on_mibi1466.blastn: 1264 MT880870_on_mibi1467.blastn: 3470 MT880870_on_mibi1468.blastn: 2372 MT880870_on_mibi1469.blastn: 2372 MT880870_on_mibi1471.blastn: 2366 MT880870_on_mibi1473.blastn: 3198 MT880870_on_mibi1474.blastn: 3198 MT880870_on_mibi1475.blastn: 3198 MT880870_on_mibi1476.blastn: 3198 MT880870_on_mibi1477.blastn: 2372 MT880870_on_mibi1478.blastn: 5918 MT880870_on_mibi1479.blastn: 3198 MT880870_on_mibi1480.blastn: 34177 MT880870_on_mibi1481.blastn: 3198 MT880870_on_mibi1482.blastn: 3198 MT880870_on_mibi1483.blastn: 3198 MT880870_on_mibi1484.blastn: 3198 MT880870_on_mibi1485.blastn: 20038 MT880870_on_mibi1486.blastn: 3198 MT880870_on_mibi1487.blastn: 28520 MT880870_on_mibi1488.blastn: 2366 MT880870_on_mibi1489.blastn: 2366 MT880870_on_mibi1490.blastn: 3198 MT880870_on_mibi1491.blastn: 3198 MT880870_on_mibi1492.blastn: 3198 MT880870_on_mibi1493.blastn: 2233 MT880870_on_mibi1494.blastn: 3198 MT880870_on_mibi1495.blastn: 3198 MT880870_on_mibi1496.blastn: 25135 MT880870_on_mibi1497.blastn: 3198 MT880870_on_mibi1498.blastn: 28012 MT880870_on_mibi1499.blastn: 3198 MT880870_on_mibi1500.blastn: 2366 MT880870_on_mibi1501.blastn: 2366 MT880870_on_mibi1502.blastn: 34177 MT880870_on_mibi1503.blastn: 2366 MT880870_on_mibi1504.blastn: 3198 MT880870_on_mibi1505.blastn: 3198 MT880870_on_mibi1506.blastn: 2372 MT880870_on_mibi2312.blastn: 3198 MT880870_on_mibi2313.blastn: 3198 MT880870_on_mibi2314.blastn: 3198 MT880870_on_mibi2315.blastn: 2366 MT880870_on_mibi2316.blastn: 3198 MT880870_on_mibi2317.blastn: 3198 MT880870_on_mibi2318.blastn: 20006 MT880870_on_mibi2319.blastn: 17544 MT880870_on_mibi2320.blastn: 3198 MT880870_on_mibi2321.blastn: 3198 MT880870_on_mibi2379.blastn: 3198 #len=36164 for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query MT880871.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./MT880871_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py MT880871_on_${sample}.blastn done MT880871_on_mibi1435.blastn: 270 MT880871_on_mibi1436.blastn: 35580 MT880871_on_mibi1437.blastn: 518 MT880871_on_mibi1438.blastn: 518 MT880871_on_mibi1439.blastn: 518 MT880871_on_mibi1440.blastn: 36682 MT880871_on_mibi1441.blastn: 519 MT880871_on_mibi1442.blastn: 514 MT880871_on_mibi1443.blastn: 0 MT880871_on_mibi1444.blastn: 0 MT880871_on_mibi1445.blastn: 518 MT880871_on_mibi1446.blastn: 518 MT880871_on_mibi1447.blastn: 518 MT880871_on_mibi1448.blastn: 518 MT880871_on_mibi1449.blastn: 518 MT880871_on_mibi1450.blastn: 518 MT880871_on_mibi1451.blastn: 0 MT880871_on_mibi1452.blastn: 30418 MT880871_on_mibi1453.blastn: 518 MT880871_on_mibi1454.blastn: 982 MT880871_on_mibi1455.blastn: 518 MT880871_on_mibi1456.blastn: 982 MT880871_on_mibi1457.blastn: 518 MT880871_on_mibi1458.blastn: 0 MT880871_on_mibi1459.blastn: 34965 MT880871_on_mibi1460.blastn: 518 MT880871_on_mibi1461.blastn: 16296 MT880871_on_mibi1462.blastn: 0 MT880871_on_mibi1463.blastn: 518 MT880871_on_mibi1464.blastn: 947 MT880871_on_mibi1465.blastn: 518 MT880871_on_mibi1466.blastn: 0 MT880871_on_mibi1467.blastn: 514 MT880871_on_mibi1468.blastn: 518 MT880871_on_mibi1469.blastn: 518 MT880871_on_mibi1471.blastn: 518 MT880871_on_mibi1473.blastn: 518 MT880871_on_mibi1474.blastn: 518 MT880871_on_mibi1475.blastn: 518 MT880871_on_mibi1476.blastn: 518 MT880871_on_mibi1477.blastn: 518 MT880871_on_mibi1478.blastn: 18607 MT880871_on_mibi1479.blastn: 518 MT880871_on_mibi1480.blastn: 36682 MT880871_on_mibi1481.blastn: 0 MT880871_on_mibi1482.blastn: 518 MT880871_on_mibi1483.blastn: 32395 MT880871_on_mibi1484.blastn: 0 MT880871_on_mibi1485.blastn: 36164 MT880871_on_mibi1486.blastn: 31072 MT880871_on_mibi1487.blastn: 36682 MT880871_on_mibi1488.blastn: 0 MT880871_on_mibi1489.blastn: 0 MT880871_on_mibi1490.blastn: 518 MT880871_on_mibi1491.blastn: 518 MT880871_on_mibi1492.blastn: 518 MT880871_on_mibi1493.blastn: 740 MT880871_on_mibi1494.blastn: 518 MT880871_on_mibi1495.blastn: 0 MT880871_on_mibi1496.blastn: 24187 MT880871_on_mibi1497.blastn: 31069 MT880871_on_mibi1498.blastn: 36682 MT880871_on_mibi1499.blastn: 518 MT880871_on_mibi1500.blastn: 0 MT880871_on_mibi1501.blastn: 0 MT880871_on_mibi1502.blastn: 36682 MT880871_on_mibi1503.blastn: 518 MT880871_on_mibi1504.blastn: 518 MT880871_on_mibi1505.blastn: 518 MT880871_on_mibi1506.blastn: 0 MT880871_on_mibi2312.blastn: 316 MT880871_on_mibi2313.blastn: 518 MT880871_on_mibi2314.blastn: 518 MT880871_on_mibi2315.blastn: 0 MT880871_on_mibi2316.blastn: 518 MT880871_on_mibi2317.blastn: 518 MT880871_on_mibi2318.blastn: 36682 MT880871_on_mibi2319.blastn: 36164 MT880871_on_mibi2320.blastn: 518 MT880871_on_mibi2321.blastn: 518 MT880871_on_mibi2379.blastn: 518 #len=147057 for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do blastn -db ../shovill/${sample}/contigs.fa -query MT880872.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880872_on_${sample}.blastn done for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do python3 analyze_blastn_res.py MT880872_on_${sample}.blastn done MT880872_on_mibi1435.blastn: 19155 MT880872_on_mibi1436.blastn: 19049 MT880872_on_mibi1437.blastn: 19049 MT880872_on_mibi1438.blastn: 19045 MT880872_on_mibi1439.blastn: 19042 MT880872_on_mibi1440.blastn: 77536 * MT880872_on_mibi1441.blastn: 19041 MT880872_on_mibi1442.blastn: 19044 MT880872_on_mibi1443.blastn: 19049 MT880872_on_mibi1444.blastn: 19049 MT880872_on_mibi1445.blastn: 19042 MT880872_on_mibi1446.blastn: 19042 MT880872_on_mibi1447.blastn: 19042 MT880872_on_mibi1448.blastn: 19049 MT880872_on_mibi1449.blastn: 19041 MT880872_on_mibi1450.blastn: 19049 MT880872_on_mibi1451.blastn: 19043 MT880872_on_mibi1452.blastn: 19047 MT880872_on_mibi1453.blastn: 19165 MT880872_on_mibi1454.blastn: 52122 * MT880872_on_mibi1455.blastn: 20996 MT880872_on_mibi1456.blastn: 52122 * MT880872_on_mibi1457.blastn: 19042 MT880872_on_mibi1458.blastn: 19048 MT880872_on_mibi1459.blastn: 77366 * MT880872_on_mibi1460.blastn: 19042 MT880872_on_mibi1461.blastn: 84536 * MT880872_on_mibi1462.blastn: 19048 MT880872_on_mibi1463.blastn: 19042 MT880872_on_mibi1464.blastn: 19049 MT880872_on_mibi1465.blastn: 19045 MT880872_on_mibi1466.blastn: 6525 MT880872_on_mibi1467.blastn: 19044 MT880872_on_mibi1468.blastn: 19037 MT880872_on_mibi1469.blastn: 19039 MT880872_on_mibi1471.blastn: 19045 MT880872_on_mibi1473.blastn: 19043 MT880872_on_mibi1474.blastn: 19052 MT880872_on_mibi1475.blastn: 19264 MT880872_on_mibi1476.blastn: 19052 MT880872_on_mibi1477.blastn: 19041 MT880872_on_mibi1478.blastn: 26492 * MT880872_on_mibi1479.blastn: 19042 MT880872_on_mibi1480.blastn: 45789 * MT880872_on_mibi1481.blastn: 19036 MT880872_on_mibi1482.blastn: 19036 MT880872_on_mibi1483.blastn: 19036 MT880872_on_mibi1484.blastn: 19045 MT880872_on_mibi1485.blastn: 52124 * MT880872_on_mibi1486.blastn: 19052 MT880872_on_mibi1487.blastn: 56847 * MT880872_on_mibi1488.blastn: 19049 MT880872_on_mibi1489.blastn: 19053 MT880872_on_mibi1490.blastn: 19045 MT880872_on_mibi1491.blastn: 19042 MT880872_on_mibi1492.blastn: 19049 MT880872_on_mibi1493.blastn: 19046 MT880872_on_mibi1494.blastn: 19049 MT880872_on_mibi1495.blastn: 19046 MT880872_on_mibi1496.blastn: 92504 * MT880872_on_mibi1497.blastn: 19052 MT880872_on_mibi1498.blastn: 58985 * MT880872_on_mibi1499.blastn: 19043 MT880872_on_mibi1500.blastn: 19052 MT880872_on_mibi1501.blastn: 19049 MT880872_on_mibi1502.blastn: 46101 * MT880872_on_mibi1503.blastn: 19044 MT880872_on_mibi1504.blastn: 19041 MT880872_on_mibi1505.blastn: 19052 MT880872_on_mibi1506.blastn: 19049 MT880872_on_mibi2312.blastn: 19045 MT880872_on_mibi2313.blastn: 19049 MT880872_on_mibi2314.blastn: 19052 MT880872_on_mibi2315.blastn: 19049 MT880872_on_mibi2316.blastn: 19052 MT880872_on_mibi2317.blastn: 19042 MT880872_on_mibi2318.blastn: 78246 * MT880872_on_mibi2319.blastn: 77512 * MT880872_on_mibi2320.blastn: 19045 MT880872_on_mibi2321.blastn: 19052 MT880872_on_mibi2379.blastn: 19156 -
yycG has only 80% similarity, but it still belongs to the gene family.
Based on the provided BLASTN result summary, determining whether it's the same gene depends on multiple factors, including sequence similarity, alignment length, and expected value (E-value). Let’s break down the results: Query ID: gi|49240382|emb|BX571856.1|:25617-27443 Subject ID: contig00005 Percentage Identity: 80.338% Alignment Length: 1836 (out of 1827 in the subject sequence) Mismatches: 349 Gap Openings: 7 E-value: 0.0 (this indicates a highly significant alignment) Bit Score: 1380 Key Points to Consider: Percentage Identity (80.34%): While 80% identity is relatively high, it’s not close enough to the 90-100% typically expected for identical or highly similar genes. However, the threshold for considering sequences as "the same gene" depends on the organism, type of gene, and biological context. For some genes or species, this might still be acceptable. Alignment Length (1836 bp): The alignment covers a large portion of both sequences (1827 in the subject sequence and 1836 in the query). This is a good indication that the sequences are largely aligned. Mismatches and Gaps: There are 349 mismatches and 7 gap openings, which suggests there are differences between the two sequences. The number of differences is significant, and the biological impact of these differences would depend on whether they occur in coding regions or regulatory elements. E-value (0.0): The E-value of 0.0 indicates that the alignment is highly significant and not due to chance. Conclusion: Based on the 80.34% identity and the number of mismatches, it is unlikely that this is exactly the same gene, but it could still be a closely related gene variant, allele, or paralog. If you are looking for an exact match, this result would not qualify. However, if you are investigating gene families or variants, this sequence could be considered closely related. Further functional or experimental validation might be necessary to determine the biological significance of these differences. -
plotTreeHeatmap
FastTree -gtr -nt variants/snippy.core_without_reference.aln > plotTreeHeatmap/snippy.core.tree library(ggtree) library(ggplot2) library(dplyr) setwd("/home/jhuang/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap/") # -- edit tree -- info <- read.csv("typing.csv", sep="\t") info$name <- info$Isolate tree <- read.tree("990_backup.tree") #cols <- c(infection='purple2', commensalism='skyblue2') #heatmapData2 <- info %>% select(Isolate, ST, SCCmec, agr.typing) heatmapData2 <- info %>% select(Isolate, ST) rn <- heatmapData2$Isolate heatmapData2$Isolate <- NULL heatmapData2 <- as.data.frame(sapply(heatmapData2, as.character)) rownames(heatmapData2) <- rn #https://bookdown.org/hneth/ds4psy/D-3-apx-colors-basics.html #"blueviolet","darkgoldenrod", "tomato","mediumpurple4","indianred", #"lightcyan3","azure3", #"magenta", #"tan","brown", heatmap.colours <- c("cornflowerblue","darkgreen","seagreen3","tan","red", "navyblue", "gold", "green","orange","pink","purple","magenta","brown", "darksalmon","chocolate4","darkkhaki", "azure3", "maroon","lightgreen", "blue","cyan", "skyblue2", "blueviolet","darkgrey") #"cornflowerblue","darkgreen","seagreen3","tan","red","green","orange","pink","brown","darkgrey", #"cornflowerblue","darkgreen","red", "darkgrey") names(heatmap.colours) <- c("2","5","7","9","14", "17","23", "35","59","73", "81","86","87","89","130","190","290", "297","325", "454","487","558","766","-") #"SCCmec_type_II(2A)","SCCmec_type_III(3A)","SCCmec_type_III(3A) and #SCCmec_type_VIII(4A)","SCCmec_type_IV(2B)","SCCmec_type_IV(2B&5)","SCCmec_type_IV(2B) and #SCCmec_type_VI(4B)","SCCmec_type_IVa(2B)","SCCmec_type_IVb(2B)","SCCmec_type_IVg(2B)","none", "I","II","III", "none") #circular #scale_color_manual(values=cols) + #geom_tippoint(aes(color=Type)) + p <- ggtree(tree, layout='circular', branch.length='none') %<+% info + geom_tiplab2(aes(label=name), offset=1, size=6.0) #, geom='text', align=TRUE, linetype=NA, hjust=1.8,check.overlap=TRUE #difference between geom_tiplab and geom_tiplab2? #+ theme(axis.text.x = element_text(angle = 30, vjust = 0.5)) + theme(axis.text = element_text(size = 20)) + scale_size(range = c(1, 20)) #font.size=10, png("ggtree.png", width=1260, height=1260) #svg("ggtree.svg", width=1260, height=1260) p dev.off() png("ggtree_and_gheatmap.png", width=1290, height=1000) #svg("ggtree_and_gheatmap.svg", width=17, height=15) gheatmap(p, heatmapData2, width=0.1,colnames_position="top", colnames_angle=90, colnames_offset_y = 0.1, hjust=0.5, font.size=6, offset = 5) + scale_fill_manual(values=heatmap.colours) + theme(legend.text = element_text(size = 14)) + theme(legend.title = element_text(size = 14)) + guides(fill=guide_legend(title=""), color = guide_legend(override.aes = list(size = 5))) dev.off() -
Response
I’ve reviewed the 85 genomes as discussed. Four samples were filtered out due to insufficient coverage: mibi2381, mibi2380, and mibi1472. Additionally, the sample mibi1470 was excluded as it could not be identified as S. epidermidis based on its MLST type. This leaves us with a total of 81 samples for downstream analysis. Attached, you’ll find: * The graphic ggtree_and_gheatmap.png * A table, presence_absence_matrix_on_genes, providing an overview of gene presence/absence and relevant typing information * FASTA files for all 81 genomes
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Kraken2 Installation and Usage Guide
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Should the inputs for GSVA be normalized or raw?
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- Risks of Rebooting into Rescue Mode
- 足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Processing Data_Michelle_RNAseq_2025
Enhanced Visualization of Gene Presence for the Selected Genes in Bongarts_S.epidermidis_HDRNA
RNA-seq Tam on CP059040.1 (Acinetobacter baumannii strain ATCC 19606)
Variant Calling for Herpes Simplex Virus 1 from Patient Sample Using Capture Probe Sequencing