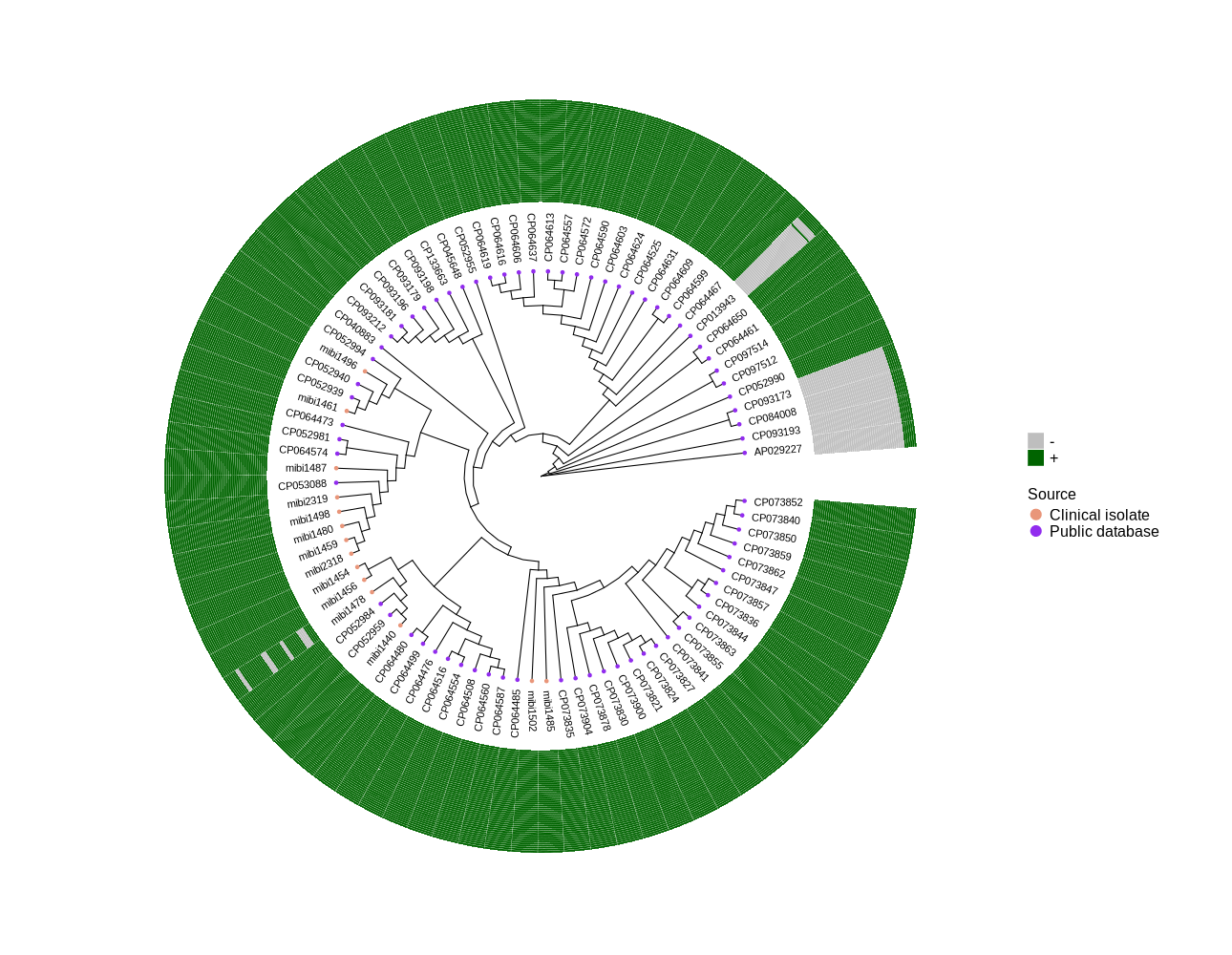
Identify all occurrences of Phage HH1 MT880870 in S. epidermidis ST2 genomes from public and clinical isolates
gene_x 0 like s 917 view s
Tags: plot, pipeline
-
install the bacto environment
mamba create -n bacto -c conda-forge -c bioconda -c defaults python=3.7 mamba activate bacto (bacto) mamba install -c conda-forge -c bioconda -c defaults trimmomatic fastqc shovill (bacto) mamba install -c conda-forge -c bioconda -c defaults mlst snippy fasttree (bacto) mamba install biopython mamba install -c conda-forge perl=5.22 mamba install -c conda-forge -c bioconda roary #------ NEED to be debugged for install roary(ERROR) --> install roary in the host env (SUCCESSFUL) ------ #ERROR (bacto) mamba install -c conda-forge -c bioconda -c defaults roary #ERROR (bacto) cpanm Bio::Roary #DEBUG NOT WORKING ! Installing Bio::Roary failed. See /home/jhuang/.cpanm/work/1736859390.2714327/build.log for details. Retry with --force to force install it. #ERROR (bacto) export PERL5LIB=$CONDA_PREFIX/lib/perl5/site_perl/5.22.0/ #DEBUG SUCCESSFUL, repaired also snippy and snippy-core sudo apt install roary (bacto) mamba install -c conda-forge -c bioconda -c defaults raxml-ng gubbins snp-sites (bacto) mamba install -c bioconda iqtree #(bacto) mamba install -c conda-forge -c bioconda -c defaults perl-bioperl #(bacto) cpanm Bio::SeqIO #(bacto) cpanm Bio::Perl #(bacto) mamba install -c conda-forge -c bioconda -c defaults biopython #(bacto) mamba install -c conda-forge -c bioconda -c defaults snakemak -
alignment of complete genomes
Alignment of Complete Genomes vs Alignment of Core Genes Alignment of Complete Genomes: Useful when all genomes are complete and high-quality. Not ideal if many genomes are incomplete or fragmented, as this can introduce noise. Alignment of Core Genes: Preferred when working with a mix of complete and draft genomes. Focuses on conserved regions, minimizing issues caused by missing or fragmented data. How to Ensure Core Gene Alignment? If your goal is to create a core gene alignment: Use a tool like Roary or Panaroo after genome annotation (e.g., with Prokka). Extract the core_gene_alignment.aln file for alignment, which represents the concatenated core genes shared across all genomes. Prepare all gff-files bpseudomallei taylorella schromogenes bcereus orhinotracheale aactinomycetemcomitans vcholerae_2 streptomyces spyogenes paeruginosa sgallolyticus bhenselae ecoli senterica_achtman_2 brachyspira leptospira_2 streptothermophilus pdamselae campylobacter_nonjejuni_3 ypseudotuberculosis_achtman_3 abaumannii_2 staphlugdunensis mhominis_3 chlamydiales cdifficile yruckeri bsubtilis sbsec koxytoca kingella tenacibaculum pgingivalis miowae neisseria bordetella_3 cbotulinum edwardsiella ureaplasma pacnes_3 mhyorhinis gallibacterium bbacilliformis sdysgalactiae shominis mcanis borrelia smaltophilia brachyspira_2 campylobacter_nonjejuni_8 psalmonis suberis achromobacter saureus mhaemolytica efaecalis cronobacter rhodococcus scanis mplutonius shewanella campylobacter_nonjejuni_7 campylobacter hparasuis listeria_2 plarvae mabscessus hcinaedi csepticum sepidermidis mhyopneumoniae mbovis_2 wolbachia vcholerae helicobacter cperfringens bcc ranatipestifer szooepidemicus msynoviae campylobacter_nonjejuni_9 pmultocida_2 arcobacter fpsychrophilum abaumannii bwashoensis mcaseolyticus liberibacter aeromonas cmaltaromaticum vvulnificus mpneumoniae klebsiella brachyspira_5 efaecium pfluorescens hsuis ssuis dnodosus sinorhizobium msciuri spneumoniae vibrio campylobacter_nonjejuni geotrichum lsalivarius tpallidum magalactiae shaemolyticus sagalactiae ecoli_achtman_4 sthermophilus bfragilis kaerogenes blicheniformis_14 vparahaemolyticus mflocculare mgallisepticum vtapetis ecloacae ppentosaceus manserisalpingitidis spseudintermedius brachyspira_4 brachyspira_3 leptospira_3 mcatarrhalis_achtman_6 oralstrep campylobacter_nonjejuni_6 pmultocida aphagocytophilum campylobacter_nonjejuni_2 otsutsugamushi diphtheria_3 hinfluenzae pputida brucella cfreundii mycobacteria_2 mgallisepticum_2 llactis_phage xfastidiosa campylobacter_nonjejuni_4 campylobacter_nonjejuni_5 leptospira python ~/Scripts/run_mlst.py complete_genome_1282_ncbi.fasta sepidermidis #73 python ~/Scripts/run_mlst.py complete_genome_1282_ena.fasta sepidermidis #73 python ~/Scripts/run_mlst.py complete_genome_1282_ena_taxon_descendants.fasta sepidermidis grep -P "\tsepidermidis\t2\t" all.mlst > ST2.mlst #73 grep -v "NZ_" ST2.mlst > ST2_.mlst cut -d'.' -f1 ST2_.mlst | sort > ST2.ids cut -d'|' -f2 ST2.mlst | sort > ST2.ids diff ST2.ids ../ena_descendants_all_ST2_mlst/ST2.ids for sample in AP029227 CP013943 CP040883 CP045648 CP052939 CP052940 CP052955 CP052959 CP052981 CP052984 CP052990 CP052994 CP053088 CP064461 CP064467 CP064473 CP064476 CP064480 CP064485 CP064499 CP064508 CP064516 CP064525 CP064554 CP064557 CP064560 CP064572 CP064574 CP064587 CP064590 CP064599 CP064603 CP064606 CP064609 CP064613 CP064616 CP064619 CP064624 CP064631 CP064637 CP064650 CP073821 CP073824 CP073827 CP073830 CP073835 CP073836 CP073840 CP073841 CP073844 CP073847 CP073850 CP073852 CP073855 CP073857 CP073859 CP073862 CP073863 CP073878 CP073900 CP073904 CP084008 CP093173 CP093179 CP093181 CP093193 CP093196 CP093198 CP093212 CP097512 CP097514 CP120425 CP133663; do cp ../genomes_by_taxid/ncbi_all_ST2_mlst/${sample}.1.fasta ./ done cd typing_complete.csv grep -P "\tsepidermidis\t2\t" typing_complete.csv > typing_ST2.csv echo -e "AP029227\nCP013943\nCP040883\nCP045648\nCP052939\nCP052940\nCP052955\nCP052959\nCP052981\nCP052984\nCP052990\nCP052994\nCP053088\nCP064461\nCP064467\nCP064473\nCP064476\nCP064480\nCP064485\nCP064499\nCP064508\nCP064516\nCP064525\nCP064554\nCP064557\nCP064560\nCP064572\nCP064574\nCP064587\nCP064590\nCP064599\nCP064603\nCP064606\nCP064609\nCP064613\nCP064616\nCP064619\nCP064624\nCP064631\nCP064637\nCP064650\nCP073821\nCP073824\nCP073827\nCP073830\nCP073835\nCP073836\nCP073840\nCP073841\nCP073844\nCP073847\nCP073850\nCP073852\nCP073855\nCP073857\nCP073859\nCP073862\nCP073863\nCP073878\nCP073900\nCP073904\nCP084008\nCP093173\nCP093179\nCP093181\nCP093193\nCP093196\nCP093198\nCP093212\nCP097512\nCP097514\nCP120425\nCP133663" > ids.txt cat ids.txt | while read id; do efetch -db nuccore -id $id -format gff3 > "${id}.gff" efetch -db nuccore -id $id -format fasta > "${id}.fasta" # Append the FASTA sequence to the GFF3 file with ##FASTA header echo "##FASTA" >> "${id}.gff" cat "${id}.fasta" >> "${id}.gff" rm "${id}.fasta" # Optionally remove the separate FASTA file done for sample in mibi1440 mibi1454 mibi1456 mibi1459 mibi1461 mibi1478 mibi1480 mibi1485 mibi1487 mibi1496 mibi1498 mibi1502 mibi2318 mibi2319; do cp ../prokka/${sample}/${sample}.gff ./ done Example with Roary: roary -f roary_ST2 -e --mafft -p 100 roary_input_ST2/*.gff #roary -p 15 -f ./roary -i 95 -cd 99 -s -e -n -v roary_.fa.gb.gff CP012351.gff3 CP003084.gff3 CP023676.gff3 -i minimum percentage identity for blastp [95] -cd FLOAT percentage of isolates a gene must be in to be core [99] -f STR output directory [.] -e create a multiFASTA alignment of core genes using PRANK -n (==--mafft) fast core gene alignment with MAFFT, use with -e -s dont split paralogs -v verbose output to STDOUT The file core_gene_alignment.aln can then be used for tree construction: iqtree2 -s core_gene_alignment.aln -m MFP -bb 1000 -nt AUTO #Alignment was printed to core_gene_alignment.aln.uniqueseq.phy #For your convenience alignment with unique sequences printed to core_gene_alignment.aln.uniqueseq.phy #Create initial parsimony tree by phylogenetic likelihood library (PLL)... 0.030 seconds #Measuring multi-threading efficiency up to 128 CPU cores #Increase to 10 rounds for branch lengths By focusing on core genes, you ensure that all genomes contribute meaningful data to the phylogenetic analysis. The file that can be viewed as the phylogenetic tree among the generated files is: core_gene_alignment.aln.treefile This file typically contains the tree in a Newick format, which is a widely used format for representing phylogenetic trees. -
Public Database and Clinical Isolates
To differentiate the two groups of genomes effectively while maintaining clarity and professionalism, you can use category names that reflect their source or origin. Here are some suggestions: Category Name Suggestions Source-based Names: Public Database and Clinical Isolates Public Genomes and Clinical Genomes Reference Genomes and Patient-derived Genomes Origin-based Names: External Sources and In-house Sequencing Global Genomes and Local Isolates Database Genomes and Clinical Strains Research Context Names: Published Genomes and Newly Sequenced Genomes Literature Genomes and Clinical Samples Archived Data and Current Study Abbreviated or Code-like Names: PUB and CLI (Public and Clinical) EXDB (External Database) and INHS (In-House Sequencing) PG (Public Genomes) and CI (Clinical Isolates) Tips for Choosing the Best Category Names Clarity: Ensure that the names are intuitive and easy for others to understand. Neutrality: Avoid names that might imply bias or inferiority (e.g., avoid "old" vs. "new"). Context Appropriateness: Consider your audience. For a clinical study, terms like "Clinical Isolates" may resonate more, while "Public Database" works better for bioinformatics research. For instance: "Database" and "Clinical" could work well in a manuscript. "External" and "In-house" might be better for internal project tracking. -
copy contigs of own clinical isolates
#extract all genes from the phages set as the column names, set the genome names from the public database and own isolates as row names. for sample in mibi1440 mibi1454 mibi1456 mibi1459 mibi1461 mibi1478 mibi1480 mibi1485 mibi1487 mibi1496 mibi1498 mibi1502 mibi2318 mibi2319; do cp ../shovill/${sample}/contigs.fa ${sample}.fasta done -
makeblastdb
#MT880870_on_mibi1435.blastn: 2366 #MT880870_on_mibi1436.blastn: 3198 #MT880870_on_mibi1437.blastn: 3198 # -lenth(MT880870)=34053 rename 's/\.1\.fasta$/.fasta/' *.1.fasta for sample in mibi1440 mibi1454 mibi1456 mibi1459 mibi1461 mibi1478 mibi1480 mibi1485 mibi1487 mibi1496 mibi1498 mibi1502 mibi2318 mibi2319 AP029227 CP013943 CP040883 CP045648 CP052939 CP052940 CP052955 CP052959 CP052981 CP052984 CP052990 CP052994 CP053088 CP064461 CP064467 CP064473 CP064476 CP064480 CP064485 CP064499 CP064508 CP064516 CP064525 CP064554 CP064557 CP064560 CP064572 CP064574 CP064587 CP064590 CP064599 CP064603 CP064606 CP064609 CP064613 CP064616 CP064619 CP064624 CP064631 CP064637 CP064650 CP073821 CP073824 CP073827 CP073830 CP073835 CP073836 CP073840 CP073841 CP073844 CP073847 CP073850 CP073852 CP073855 CP073857 CP073859 CP073862 CP073863 CP073878 CP073900 CP073904 CP084008 CP093173 CP093179 CP093181 CP093193 CP093196 CP093198 CP093212 CP097512 CP097514 CP120425 CP133663; do makeblastdb -in ${sample}.fasta -dbtype nucl done -
prepare typing_ST2_until_QPB07622.csv
samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07572.1_1" > QPB07572.fasta mkdir QPB07572 for sample in mibi1440 mibi1454 mibi1456 mibi1459 mibi1461 mibi1478 mibi1480 mibi1485 mibi1487 mibi1496 mibi1498 mibi1502 mibi2318 mibi2319 AP029227 CP013943 CP040883 CP045648 CP052939 CP052940 CP052955 CP052959 CP052981 CP052984 CP052990 CP052994 CP053088 CP064461 CP064467 CP064473 CP064476 CP064480 CP064485 CP064499 CP064508 CP064516 CP064525 CP064554 CP064557 CP064560 CP064572 CP064574 CP064587 CP064590 CP064599 CP064603 CP064606 CP064609 CP064613 CP064616 CP064619 CP064624 CP064631 CP064637 CP064650 CP073821 CP073824 CP073827 CP073830 CP073835 CP073836 CP073840 CP073841 CP073844 CP073847 CP073850 CP073852 CP073855 CP073857 CP073859 CP073862 CP073863 CP073878 CP073900 CP073904 CP084008 CP093173 CP093179 CP093181 CP093193 CP093196 CP093198 CP093212 CP097512 CP097514 CP120425 CP133663; do blastn -db ${sample}.fasta -query QPB07572.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./QPB07572/${sample}.blastn done python ~/Scripts/process_directory.py /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/QPB07572 /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/typing_ST2.csv samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07573.1_2" > QPB07573.fasta mkdir QPB07573 for sample in mibi1440 mibi1454 mibi1456 mibi1459 mibi1461 mibi1478 mibi1480 mibi1485 mibi1487 mibi1496 mibi1498 mibi1502 mibi2318 mibi2319 AP029227 CP013943 CP040883 CP045648 CP052939 CP052940 CP052955 CP052959 CP052981 CP052984 CP052990 CP052994 CP053088 CP064461 CP064467 CP064473 CP064476 CP064480 CP064485 CP064499 CP064508 CP064516 CP064525 CP064554 CP064557 CP064560 CP064572 CP064574 CP064587 CP064590 CP064599 CP064603 CP064606 CP064609 CP064613 CP064616 CP064619 CP064624 CP064631 CP064637 CP064650 CP073821 CP073824 CP073827 CP073830 CP073835 CP073836 CP073840 CP073841 CP073844 CP073847 CP073850 CP073852 CP073855 CP073857 CP073859 CP073862 CP073863 CP073878 CP073900 CP073904 CP084008 CP093173 CP093179 CP093181 CP093193 CP093196 CP093198 CP093212 CP097512 CP097514 CP120425 CP133663; do blastn -db ${sample}.fasta -query QPB07573.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./QPB07573/${sample}.blastn done python ~/Scripts/process_directory.py /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/QPB07573 /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/updated_typing_ST2.csv samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07574.1_3" > QPB07574.fasta mkdir QPB07574 for sample in mibi1440 mibi1454 mibi1456 mibi1459 mibi1461 mibi1478 mibi1480 mibi1485 mibi1487 mibi1496 mibi1498 mibi1502 mibi2318 mibi2319 AP029227 CP013943 CP040883 CP045648 CP052939 CP052940 CP052955 CP052959 CP052981 CP052984 CP052990 CP052994 CP053088 CP064461 CP064467 CP064473 CP064476 CP064480 CP064485 CP064499 CP064508 CP064516 CP064525 CP064554 CP064557 CP064560 CP064572 CP064574 CP064587 CP064590 CP064599 CP064603 CP064606 CP064609 CP064613 CP064616 CP064619 CP064624 CP064631 CP064637 CP064650 CP073821 CP073824 CP073827 CP073830 CP073835 CP073836 CP073840 CP073841 CP073844 CP073847 CP073850 CP073852 CP073855 CP073857 CP073859 CP073862 CP073863 CP073878 CP073900 CP073904 CP084008 CP093173 CP093179 CP093181 CP093193 CP093196 CP093198 CP093212 CP097512 CP097514 CP120425 CP133663; do blastn -db ${sample}.fasta -query QPB07574.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./QPB07574/${sample}.blastn done python ~/Scripts/process_directory.py /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/QPB07574 /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/updated_updated_typing_ST2.csv samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07575.1_4" > QPB07575.fasta mkdir QPB07575 for sample in mibi1440 mibi1454 mibi1456 mibi1459 mibi1461 mibi1478 mibi1480 mibi1485 mibi1487 mibi1496 mibi1498 mibi1502 mibi2318 mibi2319 AP029227 CP013943 CP040883 CP045648 CP052939 CP052940 CP052955 CP052959 CP052981 CP052984 CP052990 CP052994 CP053088 CP064461 CP064467 CP064473 CP064476 CP064480 CP064485 CP064499 CP064508 CP064516 CP064525 CP064554 CP064557 CP064560 CP064572 CP064574 CP064587 CP064590 CP064599 CP064603 CP064606 CP064609 CP064613 CP064616 CP064619 CP064624 CP064631 CP064637 CP064650 CP073821 CP073824 CP073827 CP073830 CP073835 CP073836 CP073840 CP073841 CP073844 CP073847 CP073850 CP073852 CP073855 CP073857 CP073859 CP073862 CP073863 CP073878 CP073900 CP073904 CP084008 CP093173 CP093179 CP093181 CP093193 CP093196 CP093198 CP093212 CP097512 CP097514 CP120425 CP133663; do blastn -db ${sample}.fasta -query QPB07575.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./QPB07575/${sample}.blastn done python ~/Scripts/process_directory.py /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/QPB07575 /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/updated_updated_updated_typing_ST2.csv samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07576.1_5" > QPB07576.fasta mkdir QPB07576 for sample in mibi1440 mibi1454 mibi1456 mibi1459 mibi1461 mibi1478 mibi1480 mibi1485 mibi1487 mibi1496 mibi1498 mibi1502 mibi2318 mibi2319 AP029227 CP013943 CP040883 CP045648 CP052939 CP052940 CP052955 CP052959 CP052981 CP052984 CP052990 CP052994 CP053088 CP064461 CP064467 CP064473 CP064476 CP064480 CP064485 CP064499 CP064508 CP064516 CP064525 CP064554 CP064557 CP064560 CP064572 CP064574 CP064587 CP064590 CP064599 CP064603 CP064606 CP064609 CP064613 CP064616 CP064619 CP064624 CP064631 CP064637 CP064650 CP073821 CP073824 CP073827 CP073830 CP073835 CP073836 CP073840 CP073841 CP073844 CP073847 CP073850 CP073852 CP073855 CP073857 CP073859 CP073862 CP073863 CP073878 CP073900 CP073904 CP084008 CP093173 CP093179 CP093181 CP093193 CP093196 CP093198 CP093212 CP097512 CP097514 CP120425 CP133663; do blastn -db ${sample}.fasta -query QPB07576.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./QPB07576/${sample}.blastn done python ~/Scripts/process_directory.py /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/QPB07576 /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/updated_updated_updated_updated_typing_ST2.csv samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07577.1_6" > QPB07577.fasta mkdir QPB07577 for sample in mibi1440 mibi1454 mibi1456 mibi1459 mibi1461 mibi1478 mibi1480 mibi1485 mibi1487 mibi1496 mibi1498 mibi1502 mibi2318 mibi2319 AP029227 CP013943 CP040883 CP045648 CP052939 CP052940 CP052955 CP052959 CP052981 CP052984 CP052990 CP052994 CP053088 CP064461 CP064467 CP064473 CP064476 CP064480 CP064485 CP064499 CP064508 CP064516 CP064525 CP064554 CP064557 CP064560 CP064572 CP064574 CP064587 CP064590 CP064599 CP064603 CP064606 CP064609 CP064613 CP064616 CP064619 CP064624 CP064631 CP064637 CP064650 CP073821 CP073824 CP073827 CP073830 CP073835 CP073836 CP073840 CP073841 CP073844 CP073847 CP073850 CP073852 CP073855 CP073857 CP073859 CP073862 CP073863 CP073878 CP073900 CP073904 CP084008 CP093173 CP093179 CP093181 CP093193 CP093196 CP093198 CP093212 CP097512 CP097514 CP120425 CP133663; do blastn -db ${sample}.fasta -query QPB07577.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./QPB07577/${sample}.blastn done python ~/Scripts/process_directory.py /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/QPB07577 /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/updated_updated_updated_updated_updated_typing_ST2.csv samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07578.1_7" > QPB07578.fasta mkdir QPB07578 for sample in mibi1440 mibi1454 mibi1456 mibi1459 mibi1461 mibi1478 mibi1480 mibi1485 mibi1487 mibi1496 mibi1498 mibi1502 mibi2318 mibi2319 AP029227 CP013943 CP040883 CP045648 CP052939 CP052940 CP052955 CP052959 CP052981 CP052984 CP052990 CP052994 CP053088 CP064461 CP064467 CP064473 CP064476 CP064480 CP064485 CP064499 CP064508 CP064516 CP064525 CP064554 CP064557 CP064560 CP064572 CP064574 CP064587 CP064590 CP064599 CP064603 CP064606 CP064609 CP064613 CP064616 CP064619 CP064624 CP064631 CP064637 CP064650 CP073821 CP073824 CP073827 CP073830 CP073835 CP073836 CP073840 CP073841 CP073844 CP073847 CP073850 CP073852 CP073855 CP073857 CP073859 CP073862 CP073863 CP073878 CP073900 CP073904 CP084008 CP093173 CP093179 CP093181 CP093193 CP093196 CP093198 CP093212 CP097512 CP097514 CP120425 CP133663; do blastn -db ${sample}.fasta -query QPB07578.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./QPB07578/${sample}.blastn done python ~/Scripts/process_directory.py /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/QPB07578 /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/updated_updated_updated_updated_updated_updated_typing_ST2.csv samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07579.1_8" > QPB07579.fasta mkdir QPB07579 for sample in mibi1440 mibi1454 mibi1456 mibi1459 mibi1461 mibi1478 mibi1480 mibi1485 mibi1487 mibi1496 mibi1498 mibi1502 mibi2318 mibi2319 AP029227 CP013943 CP040883 CP045648 CP052939 CP052940 CP052955 CP052959 CP052981 CP052984 CP052990 CP052994 CP053088 CP064461 CP064467 CP064473 CP064476 CP064480 CP064485 CP064499 CP064508 CP064516 CP064525 CP064554 CP064557 CP064560 CP064572 CP064574 CP064587 CP064590 CP064599 CP064603 CP064606 CP064609 CP064613 CP064616 CP064619 CP064624 CP064631 CP064637 CP064650 CP073821 CP073824 CP073827 CP073830 CP073835 CP073836 CP073840 CP073841 CP073844 CP073847 CP073850 CP073852 CP073855 CP073857 CP073859 CP073862 CP073863 CP073878 CP073900 CP073904 CP084008 CP093173 CP093179 CP093181 CP093193 CP093196 CP093198 CP093212 CP097512 CP097514 CP120425 CP133663; do blastn -db ${sample}.fasta -query QPB07579.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./QPB07579/${sample}.blastn done python ~/Scripts/process_directory.py /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/QPB07579 /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/updated_updated_updated_updated_updated_updated_updated_typing_ST2.csv grep ">" MT880870.fasta > temp cut -d' ' -f1 temp > temp1 # region="lcl|MT880870.1_cds_${id}.1_$((i - 78))" # Adjust index calculation for region # echo "samtools faidx MT880870.fasta \"$region\" > ${id}.fasta" samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07580.1_9" > QPB07580.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07581.1_10" > QPB07581.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07582.1_11" > QPB07582.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07583.1_12" > QPB07583.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07584.1_13" > QPB07584.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07585.1_14" > QPB07585.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07586.1_15" > QPB07586.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07587.1_16" > QPB07587.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07588.1_17" > QPB07588.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07589.1_18" > QPB07589.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07590.1_19" > QPB07590.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07591.1_20" > QPB07591.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07592.1_21" > QPB07592.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07593.1_22" > QPB07593.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07594.1_23" > QPB07594.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07595.1_24" > QPB07595.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07596.1_25" > QPB07596.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07597.1_26" > QPB07597.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07598.1_27" > QPB07598.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07599.1_28" > QPB07599.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07600.1_29" > QPB07600.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07601.1_30" > QPB07601.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07602.1_31" > QPB07602.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07603.1_32" > QPB07603.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07604.1_33" > QPB07604.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07605.1_34" > QPB07605.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07606.1_35" > QPB07606.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07607.1_36" > QPB07607.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07608.1_37" > QPB07608.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07609.1_38" > QPB07609.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07610.1_39" > QPB07610.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07611.1_40" > QPB07611.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07612.1_41" > QPB07612.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07613.1_42" > QPB07613.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07614.1_43" > QPB07614.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07615.1_44" > QPB07615.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07616.1_45" > QPB07616.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07617.1_46" > QPB07617.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07618.1_47" > QPB07618.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07619.1_48" > QPB07619.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07620.1_49" > QPB07620.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07621.1_50" > QPB07621.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07622.1_51" > QPB07622.fasta for i in {580..580}; do id=$(printf "QPB07%03d" "$i") echo "mkdir ${id}" echo "for sample in mibi1440 mibi1454 mibi1456 mibi1459 mibi1461 mibi1478 mibi1480 mibi1485 mibi1487 mibi1496 mibi1498 mibi1502 mibi2318 mibi2319 AP029227 CP013943 CP040883 CP045648 CP052939 CP052940 CP052955 CP052959 CP052981 CP052984 CP052990 CP052994 CP053088 CP064461 CP064467 CP064473 CP064476 CP064480 CP064485 CP064499 CP064508 CP064516 CP064525 CP064554 CP064557 CP064560 CP064572 CP064574 CP064587 CP064590 CP064599 CP064603 CP064606 CP064609 CP064613 CP064616 CP064619 CP064624 CP064631 CP064637 CP064650 CP073821 CP073824 CP073827 CP073830 CP073835 CP073836 CP073840 CP073841 CP073844 CP073847 CP073850 CP073852 CP073855 CP073857 CP073859 CP073862 CP073863 CP073878 CP073900 CP073904 CP084008 CP093173 CP093179 CP093181 CP093193 CP093196 CP093198 CP093212 CP097512 CP097514 CP120425 CP133663; do" echo "blastn -db \${sample}.fasta -query ${id}.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./${id}/\${sample}.blastn" echo "done" done python ~/Scripts/process_directory.py /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/QPB07580 /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/updated_updated_updated_updated_updated_updated_updated_updated_typing_ST2.csv /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/typing_ST2_until_QPB07580.csv for i in {581..622}; do id_1=$(printf "QPB07%03d" "$((i-1))") id=$(printf "QPB07%03d" "$i") echo "mkdir ${id}" echo "for sample in mibi1440 mibi1454 mibi1456 mibi1459 mibi1461 mibi1478 mibi1480 mibi1485 mibi1487 mibi1496 mibi1498 mibi1502 mibi2318 mibi2319 AP029227 CP013943 CP040883 CP045648 CP052939 CP052940 CP052955 CP052959 CP052981 CP052984 CP052990 CP052994 CP053088 CP064461 CP064467 CP064473 CP064476 CP064480 CP064485 CP064499 CP064508 CP064516 CP064525 CP064554 CP064557 CP064560 CP064572 CP064574 CP064587 CP064590 CP064599 CP064603 CP064606 CP064609 CP064613 CP064616 CP064619 CP064624 CP064631 CP064637 CP064650 CP073821 CP073824 CP073827 CP073830 CP073835 CP073836 CP073840 CP073841 CP073844 CP073847 CP073850 CP073852 CP073855 CP073857 CP073859 CP073862 CP073863 CP073878 CP073900 CP073904 CP084008 CP093173 CP093179 CP093181 CP093193 CP093196 CP093198 CP093212 CP097512 CP097514 CP120425 CP133663; do" echo "blastn -db \${sample}.fasta -query ${id}.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./${id}/\${sample}.blastn" echo "done" echo "python ~/Scripts/process_directory.py /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/${id} /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/typing_ST2_until_${id_1}.csv /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/typing_ST2_until_${id}.csv" done cp typing_ST2_until_QPB07622.csv ../plotTreeHeatmap_ST2/typing_ST2.csv -
plot tree heatmap
library(ggtree) library(ggplot2) library(dplyr) setwd("/home/jhuang/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_ST2/") # -- edit tree -- #https://icytree.org/ #0.000780 info <- read.csv("typing_ST2.csv", sep="\t") info$name <- info$Isolate tree <- read.tree("core_gene_alignment.aln.treefile") cols <- c("Public database"='purple2', "Clinical isolate"='darksalmon') #purple2 skyblue2 heatmapData2 <- info %>% select(Isolate, QPB07572, QPB07573, QPB07574, QPB07575, QPB07576, QPB07577, QPB07578, QPB07579, QPB07580, QPB07581, QPB07582, QPB07583, QPB07584, QPB07585, QPB07586, QPB07587, QPB07588, QPB07589, QPB07590, QPB07591, QPB07592, QPB07593, QPB07594, QPB07595, QPB07596, QPB07597, QPB07598, QPB07599, QPB07600, QPB07601, QPB07602, QPB07603, QPB07604, QPB07605, QPB07606, QPB07607, QPB07608, QPB07609, QPB07610, QPB07611, QPB07612, QPB07613, QPB07614, QPB07615, QPB07616, QPB07617, QPB07618, QPB07619, QPB07620, QPB07621, QPB07622) #ST, rn <- heatmapData2$Isolate heatmapData2$Isolate <- NULL heatmapData2 <- as.data.frame(sapply(heatmapData2, as.character)) rownames(heatmapData2) <- rn heatmap.colours <- c("darkgreen","grey") names(heatmap.colours) <- c("+","-") #mydat$Regulation <- factor(mydat$Regulation, levels=c("up","down")) #circular p <- ggtree(tree, layout='circular', branch.length='none') %<+% info + geom_tippoint(aes(color=Source)) + scale_color_manual(values=cols) + geom_tiplab2(aes(label=name), offset=1) png("ggtree.png", width=1260, height=1260) #svg("ggtree.svg", width=1260, height=1260) p dev.off() #png("ggtree_and_gheatmap.png", width=1290, height=1000) #svg("ggtree_and_gheatmap.svg", width=17, height=15) #gheatmap(p, heatmapData2, width=0.1, colnames_position="top", colnames_angle=90, colnames_offset_y = 0.1, hjust=0.5, font.size=4, offset = 5) + scale_fill_manual(values=heatmap.colours) + theme(legend.text = element_text(size = 14)) + theme(legend.title = element_text(size = 14)) + guides(fill=guide_legend(title=""), color = guide_legend(override.aes = list(size = 5))) png("ggtree_and_gheatmap.png", width=1290, height=1000) gheatmap(p, heatmapData2, width=0.5, colnames_position="top", colnames_angle=90, colnames_offset_y = 0.1, hjust=0.5, font.size=0, offset = 8) + scale_fill_manual(values=heatmap.colours) + theme(legend.text = element_text(size = 16)) + theme(legend.title = element_text(size = 16)) + guides(fill=guide_legend(title=""), color = guide_legend(override.aes = list(size = 5))) dev.off()
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Kraken2 Installation and Usage Guide
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Should the inputs for GSVA be normalized or raw?
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- Risks of Rebooting into Rescue Mode
- 足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Enhanced Visualization of Gene Presence for the Selected Genes in Bongarts_S.epidermidis_HDRNA