Comprehensive smallRNA-7 profiling using exceRpt pipeline with full reference databases
gene_x 0 like s 632 view s
Tags: pipeline
TODO_1: Update the image
-
Input data
mkdir ~/DATA/Data_Ute/Data_Ute_smallRNA_7/raw_data cd raw_data cp ~/DATA/Data_Ute/Data_Ute_smallRNA_7/231016_NB501882_0435_AHG7HMBGXV/nf930/01_0505_WaGa_wt_EV_RNA_S1_R1_001.fastq.gz 0505_WaGa_wt.fastq.gz cp ~/DATA/Data_Ute/Data_Ute_smallRNA_7/231016_NB501882_0435_AHG7HMBGXV/nf931/02_0505_WaGa_sT_DMSO_EV_RNA_S2_R1_001.fastq.gz 0505_WaGa_sT_DMSO.fastq.gz cp ~/DATA/Data_Ute/Data_Ute_smallRNA_7/231016_NB501882_0435_AHG7HMBGXV/nf932/03_0505_WaGa_sT_Dox_EV_RNA_S3_R1_001.fastq.gz 0505_WaGa_sT_Dox.fastq.gz cp ~/DATA/Data_Ute/Data_Ute_smallRNA_7/231016_NB501882_0435_AHG7HMBGXV/nf933/04_0505_WaGa_scr_DMSO_EV_RNA_S4_R1_001.fastq.gz 0505_WaGa_scr_DMSO.fastq.gz cp ~/DATA/Data_Ute/Data_Ute_smallRNA_7/231016_NB501882_0435_AHG7HMBGXV/nf934/05_0505_WaGa_scr_Dox_EV_RNA_S5_R1_001.fastq.gz 0505_WaGa_scr_Dox.fastq.gz cp ~/DATA/Data_Ute/Data_Ute_smallRNA_7/231016_NB501882_0435_AHG7HMBGXV/nf935/06_1905_WaGa_wt_EV_RNA_S6_R1_001.fastq.gz 1905_WaGa_wt.fastq.gz cp ~/DATA/Data_Ute/Data_Ute_smallRNA_7/231016_NB501882_0435_AHG7HMBGXV/nf936/07_1905_WaGa_sT_DMSO_EV_RNA_S7_R1_001.fastq.gz 1905_WaGa_sT_DMSO.fastq.gz cp ~/DATA/Data_Ute/Data_Ute_smallRNA_7/231016_NB501882_0435_AHG7HMBGXV/nf937/08_1905_WaGa_sT_Dox_EV_RNA_S8_R1_001.fastq.gz 1905_WaGa_sT_Dox.fastq.gz cp ~/DATA/Data_Ute/Data_Ute_smallRNA_7/231016_NB501882_0435_AHG7HMBGXV/nf938/09_1905_WaGa_scr_DMSO_EV_RNA_S9_R1_001.fastq.gz 1905_WaGa_scr_DMSO.fastq.gz cp ~/DATA/Data_Ute/Data_Ute_smallRNA_7/231016_NB501882_0435_AHG7HMBGXV/nf939/10_1905_WaGa_scr_Dox_EV_RNA_S10_R1_001.fastq.gz 1905_WaGa_scr_Dox.fastq.gz cp ~/DATA/Data_Ute/Data_Ute_smallRNA_7/231016_NB501882_0435_AHG7HMBGXV/nf940/11_control_MKL1_S11_R1_001.fastq.gz control_MKL1.fastq.gz cp ~/DATA/Data_Ute/Data_Ute_smallRNA_7/231016_NB501882_0435_AHG7HMBGXV/nf941/12_control_WaGa_S12_R1_001.fastq.gz control_WaGa.fastq.gz #END -
Run cutadapt
some common adapter sequences from different kits for reference: - TruSeq Small RNA (Illumina): TGGAATTCTCGGGTGCCAAGG - Small RNA Kits V1 (Illumina): TCGTATGCCGTCTTCTGCTTGT - Small RNA Kits V1.5 (Illumina): ATCTCGTATGCCGTCTTCTGCTTG - NEXTflex Small RNA Sequencing Kit v3 for Illumina Platforms (Bioo Scientific): TGGAATTCTCGGGTGCCAAGG - LEXOGEN Small RNA-Seq Library Prep Kit (Illumina): TGGAATTCTCGGGTGCCAAGGAACTCCAGTCAC * mkdir trimmed; cd trimmed for sample in 0505_WaGa_wt 0505_WaGa_sT_DMSO 0505_WaGa_sT_Dox 0505_WaGa_scr_DMSO 0505_WaGa_scr_Dox 1905_WaGa_wt 1905_WaGa_sT_DMSO 1905_WaGa_sT_Dox 1905_WaGa_scr_DMSO 1905_WaGa_scr_Dox control_MKL1 control_WaGa; do cutadapt -a TGGAATTCTCGGGTGCCAAGGAACTCCAGTCAC -q 20 -o ${sample}_cutadapted.fastq.gz --minimum-length 5 --trim-n ../raw_data/${sample}.fastq.gz >> LOG done # -- check if it is necessary to remove adapter from 5'-end -- (Option_1) cutadapt -g TGGAATTCTCGGGTGCCAAGGAACTCCAGTCAC -o /dev/null --report=minimal 0505_WaGa_wt_cutadapted.fastq.gz --> The trimming statistics in the output will show how often 5'-end adapters were removed. (Option 2) zcat your_sample.fastq.gz | grep 'TGGAATTCTCGGGTGCCAAGGAACTCCAGTCAC' | head -n 20 (Option 3) fastqc your_sample.fastq.gz #Open the generated HTML report and check: # The "Overrepresented sequences" section for adapter sequences. # The "Per base sequence content" plot to see if there are unexpected sequences at the start of reads. #(If check results shows both ends contain adapter) cutadapt -g TGGAATTCTCGGGTGCCAAGGAACTCCAGTCAC -a TGGAATTCTCGGGTGCCAAGGAACTCCAGTCAC -q 20 --minimum-length 10 -o ${sample}_trimmed.fastq.gz ${sample}.fastq.gz >> LOG2 # -g → Trims 5'-end adapters # -a → Trims 3'-end adapters; -a TGGAATTCTCGGGTGCCAAGGAACTCCAGTCAC → Specifies the adapter sequence to be removed from the 3' end of the reads. The sequence provided is common in RNA-seq libraries (e.g., Illumina small RNA sequencing). # -q 20 → Performs quality trimming at both read ends, removing bases with a Phred quality score below 20. -
Install exceRpt (https://github.gersteinlab.org/exceRpt/)
docker pull rkitchen/excerpt mkdir MyexceRptDatabase cd /mnt/nvme0n1p1/MyexceRptDatabase wget http://org.gersteinlab.excerpt.s3-website-us-east-1.amazonaws.com/exceRptDB_v4_hg38_lowmem.tgz tar -xvf exceRptDB_v4_hg38_lowmem.tgz #http://org.gersteinlab.excerpt.s3-website-us-east-1.amazonaws.com/exceRptDB_v4_hg19_lowmem.tgz #http://org.gersteinlab.excerpt.s3-website-us-east-1.amazonaws.com/exceRptDB_v4_hg38_lowmem.tgz #http://org.gersteinlab.excerpt.s3-website-us-east-1.amazonaws.com/exceRptDB_v4_mm10_lowmem.tgz wget http://org.gersteinlab.excerpt.s3-website-us-east-1.amazonaws.com/exceRptDB_v4_EXOmiRNArRNA.tgz tar -xvf exceRptDB_v4_EXOmiRNArRNA.tgz wget http://org.gersteinlab.excerpt.s3-website-us-east-1.amazonaws.com/exceRptDB_v4_EXOGenomes.tgz tar -xvf exceRptDB_v4_EXOGenomes.tgz -
Run exceRpt
#[COMPLETE_DB] docker run -v /mnt/nvme0n1p1/MyInputSample:/exceRptInput \ -v /mnt/nvme0n1p1/MyResults:/exceRptOutput \ -v /mnt/nvme0n1p1/MyexceRptDatabase:/exceRpt_DB \ -t rkitchen/excerpt \ INPUT_FILE_PATH=/exceRptInput/0505_WaGa_wt_cutadapted.fastq.gz \ MAIN_ORGANISM_GENOME_ID=hg38 \ N_THREADS=50 \ JAVA_RAM='800G' #[SMALL_DB] docker run -v /mnt/nvme0n1p1/MyInputSample:/exceRptInput \ -v /mnt/nvme0n1p1/MyResults:/exceRptOutput \ -v /mnt/nvme0n1p1/MyexceRptDatabase/hg38:/exceRpt_DB/hg38 \ -t rkitchen/excerpt \ INPUT_FILE_PATH=/exceRptInput/${sample}_cutadapted.fastq.gz N_THREADS=50 \ JAVA_RAM='800G' #[REAL_RUNNING_SMALL_DB] mkdir results for sample in 0505_WaGa_wt 0505_WaGa_sT_DMSO 0505_WaGa_sT_Dox 0505_WaGa_scr_DMSO 0505_WaGa_scr_Dox 1905_WaGa_wt 1905_WaGa_sT_DMSO 1905_WaGa_sT_Dox 1905_WaGa_scr_DMSO 1905_WaGa_scr_Dox control_MKL1 control_WaGa; do docker run -v ~/DATA/Data_Ute/Data_Ute_smallRNA_7/trimmed:/exceRptInput \ -v ~/DATA/Data_Ute/Data_Ute_smallRNA_7/results:/exceRptOutput \ -v /mnt/nvme0n1p1/MyexceRptDatabase/hg38:/exceRpt_DB/hg38 \ -t rkitchen/excerpt \ INPUT_FILE_PATH=/exceRptInput/${sample}_cutadapted.fastq.gz MAIN_ORGANISM_GENOME_ID=hg38 N_THREADS=50 JAVA_RAM='200G' done mkdir results_exo2 for sample in 0505_WaGa_wt; do for sample in 0505_WaGa_sT_DMSO 0505_WaGa_sT_Dox 0505_WaGa_scr_DMSO 0505_WaGa_scr_Dox 1905_WaGa_wt 1905_WaGa_sT_DMSO 1905_WaGa_sT_Dox 1905_WaGa_scr_DMSO 1905_WaGa_scr_Dox control_MKL1 control_WaGa; do docker run -v ~/DATA/Data_Ute/Data_Ute_smallRNA_7/trimmed:/exceRptInput \ -v ~/DATA/Data_Ute/Data_Ute_smallRNA_7/results_exo2:/exceRptOutput \ -v /mnt/nvme0n1p1/MyexceRptDatabase/hg38:/exceRpt_DB/gh38 \ -v /mnt/nvme0n1p1/MyexceRptDatabase/miRBase:/exceRpt_DB/miRBase \ -v /mnt/nvme0n1p1/MyexceRptDatabase/NCBI_taxonomy_taxdump:/exceRpt_DB/NCBI_taxonomy_taxdump \ -v /mnt/nvme0n1p1/MyexceRptDatabase/Genomes_BacteriaFungiMammalPlantProtistVirus:/exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus \ -v /mnt/nvme0n1p1/MyexceRptDatabase/ribosomeDatabase:/exceRpt_DB/ribosomeDatabase \ -t rkitchen/excerpt \ INPUT_FILE_PATH=/exceRptInput/${sample}_cutadapted.fastq.gz MAIN_ORGANISM_GENOME_ID=hg38 N_THREADS=50 JAVA_RAM='200G' MAP_EXOGENOUS=on done #DEBUG_1 for ERROR: could not find adapters at path /exceRpt_DB/adapters/adapters.fa #The /exceRpt_DB/adapters/adapters.fa in the Docker environment will be overwritten when assigning a new directory as /exceRpt_DB. Therefore, we should create a new adapters.fa file in the new database environment jhuang@WS-2290C:/mnt/nvme0n1p1/MyexceRptDatabase$ cp -r ../exceRpt/exceRpt_coreDB/* ./ #DEBUG_2 for EXITING because of fatal input ERROR: could not open user-defined parameters file /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in #jhuang@WS-2290C:/mnt/nvme0n1p1/MyexceRptDatabase$ cp STAR_Parameters_Exogenous.in Genomes_BacteriaFungiMammalPlantProtistVirus/ #Debugging Tips # Verify Database Structure and Ensure your mounted /exceRpt_DB contains: # /exceRpt_DB # ├── hg38/ # Endogenous # ├── NCBI_taxonomy_taxdump/ # Taxonomy # └── Genomes_BacteriaFungi.../ # Exogenous references # Check Intermediate Files # Confirm that the endogenous step generates the expected input for exogenous processing (e.g., exogenous_alignments.sam). mkdir results_g results_exo4 results_exo5 docker run -v ~/DATA/Data_Ute/Data_Ute_smallRNA_7/results_exo4:/exceRptOutput \ -v /mnt/nvme0n1p1/MyexceRptDatabase:/exceRpt_DB \ -t rkitchen/excerpt \ INPUT_FILE_PATH=/exceRptInput/testData_human.fastq.gz MAIN_ORGANISM_GENOME_ID=hg38 N_THREADS=50 JAVA_RAM='200G' MAP_EXOGENOUS=on #NOTE that rkitchen/excerpt refers to exceRpt_shortRNA (bash script): The extra-cellular RNA processing toolkit (exceRpt) optimised for smallRNA analysis; This pipeline processes a single smallRNA sequence file from a single sample #TODO_3: how to call exceRpt_longRNA: The extra-cellular RNA processing toolkit (exceRpt) optimised for longRNA analysis; This pipeline processes a single longRNA sequence file from a single sample. # docker inspect rkitchen/excerpt:latest; docker history rkitchen/excerpt:latest; docker history --no-trunc rkitchen/excerpt:latest # "Entrypoint": [ # "make", # "-f", # "/exceRpt_bin/exceRpt_smallRNA", # "EXE_DIR=/exceRpt_bin", # "DATABASE_PATH=/exceRpt_DB", # "JAVA_EXE=java", # "OUTPUT_DIR=/exceRptOutput", # "MAP_EXOGENOUS=off", # "N_THREADS=4" # ] #[REAL_RUNNING_COMPLETE_DB] #NOTE that if not renamed in the input files, then have to RENAME all files recursively by removing "_cutadapted.fastq" in all names in _CORE_RESULTS_v4.6.3.tgz (first unzip, removing, then zip, mv to ../results_g). cd trimmed for file in *_cutadapted.fastq.gz; do echo "mv \"$file\" \"${file/_cutadapted.fastq/}\"" done mkdir results_exo5 for sample in 0505_WaGa_wt 0505_WaGa_sT_DMSO 0505_WaGa_sT_Dox 0505_WaGa_scr_DMSO 0505_WaGa_scr_Dox 1905_WaGa_wt 1905_WaGa_sT_DMSO 1905_WaGa_sT_Dox 1905_WaGa_scr_DMSO 1905_WaGa_scr_Dox control_MKL1 control_WaGa; do docker run -v ~/DATA/Data_Ute/Data_Ute_smallRNA_7/trimmed:/exceRptInput \ -v ~/DATA/Data_Ute/Data_Ute_smallRNA_7/results_exo5:/exceRptOutput \ -v /mnt/nvme0n1p1/MyexceRptDatabase:/exceRpt_DB \ -t rkitchen/excerpt \ INPUT_FILE_PATH=/exceRptInput/${sample}.gz MAIN_ORGANISM_GENOME_ID=hg38 N_THREADS=50 JAVA_RAM='200G' MAP_EXOGENOUS=on done #The running process: https://github.com/gersteinlab/exceRpt/blob/master/exceRpt_smallRNA (bash script) in docker, then call java scripts https://github.com/gersteinlab/exceRpt/blob/master/exceRpt_Tools/main/ExceRpt_Tools.java, ProcessEndogenousAlignments.java and ProcessExogenousAlignments.java. #NOTE that in exceRpt_smallRNA.sh ## Choose what kind of EXOGENOUS alignments to attempt: ## - off : none ## - miRNA : map only to exogenous miRNAs in miRbase ## - on : map to exogenous miRNAs in miRbase AND the genomes of all sequenced species in ensembl/NCBI #Most of the Docker command is loading directories on your machine (the -v parameters) so that exceRpt can read from or write to them. The directory to the left of each : can obviously be whatever you want, but it is important to make sure the right side of each : is written as above or exceRpt will not be able to find/write the data it needs. -
Processing exceRpt output from multiple samples
Also provided is a script to combine output from multiple samples run through the exceRpt pipeline. The script (mergePipelineRuns.R) will take as input a directory containing 1 or more subdirectories or zipfiles containing output from the makefile above. In this way, results from 1 or more smallRNA-seq samples can be combined, several QC plots are generated, and the read-counts are normalised ready for downstream analysis by clustering and/or differential expression.
Installation
This script is comparatively much simpler to install. Once the R software (http://cran.r-project.org/) is set up on your system the script should automatically identify and install all required dependencies. Again, this script is available on the Genboree Workbench (www.genboree.org) and is also free for academic use.Using the script: On the command line
mamba activate r_env jhuang@WS-2290C:/mnt/nvme0n1p1/exceRpt-master$ Rscript mergePipelineRuns.R /home/jhuang/DATA/Data_Ute/Data_Ute_smallRNA_7/MyResults/ #OBSERVE the env of R: ~/mambaforge/envs/r_env/lib/R/library #which R: /home/jhuang/mambaforge/envs/r_env/bin/R #The env is nothing to do with "sudo chmod -R 777 /usr/lib/R/site-library" #ERROR: MyResults is not writable --> DEBUG: sudo chown -R jhuang:jhuang MyResults MyResults2 results results2-- COUNTINE HERE after docker running --> Using the script: Interactively in R
#Alternatively in an interactive R session, the merge can be performed using the following two commands: mkdir summaries_g summaries_exo4 summaries_exo5 (r_env) jhuang@WS-2290C:~/DATA/Data_Ute/Data_Ute_smallRNA_7/exceRpt-master$ R #WARNING: need to reload the R-script after each change of the script. source("mergePipelineRuns_functions.R") # -- DEBUG freetype-error -- # #sudo apt-get install libfreetype6-dev # mamba activate r_env # mamba install -c conda-forge --force-reinstall freetype fontconfig pkg-config # library(systemfonts) # system_fonts() # Should return font list without errors getwd() [1] "/media/jhuang/Elements/Data_Ute/Data_Ute_smallRNA_7/exceRpt-master" processSamplesInDir("../results_g/", "../summaries_g") processSamplesInDir("../results_exo4/", "../summaries_exo4") processSamplesInDir("../results_exo5/", "../summaries_exo5") #~/Tools/csv2xls-0.4/csv_to_xls.py exceRpt_miRNA_ReadsPerMillion.txt exceRpt_tRNA_ReadsPerMillion.txt exceRpt_piRNA_ReadsPerMillion.txt -d$'\t' -o exceRpt_results_detailed.xlsScript output
Several files are output by the script in the location of the input exceRpt results (or somewhere else if explicitly specified). All output files are prefixed with ‘exceRpt_’ and contain a variety of information regarding all samples input: File Name Description QC data: exceRpt_DiagnosticPlots.pdf All diagnostic plots automatically generated by the merge script exceRpt_readMappingSummary.txt Read-alignment summary including total counts for each library exceRpt_ReadLengths.txt Read-lengths (after 3’ adapters/barcodes are removed) Raw transcriptome quantifications: exceRpt_miRNA_ReadCounts.txt miRNA read-counts quantifications exceRpt_tRNA_ReadCounts.txt tRNA read-counts quantifications exceRpt_piRNA_ReadCounts.txt piRNA read-counts quantifications exceRpt_gencode_ReadCounts.txt gencode read-counts quantifications exceRpt_circularRNA_ReadCounts.txt circularRNA read-count quantifications Normalised transcriptome quantifications: exceRpt_miRNA_ReadsPerMillion.txt miRNA RPM quantifications exceRpt_tRNA_ReadsPerMillion.txt tRNA RPM quantifications exceRpt_piRNA_ReadsPerMillion.txt piRNA RPM quantifications exceRpt_gencode_ReadsPerMillion.txt gencode RPM quantifications exceRpt_circularRNA_ReadsPerMillion.txt circularRNA RPM quantifications R objects: exceRpt_smallRNAQuants_ReadCounts.RData All raw data (binary R object) exceRpt_smallRNAQuants_ReadsPerMillion.RData All normalised data (binary R object) -
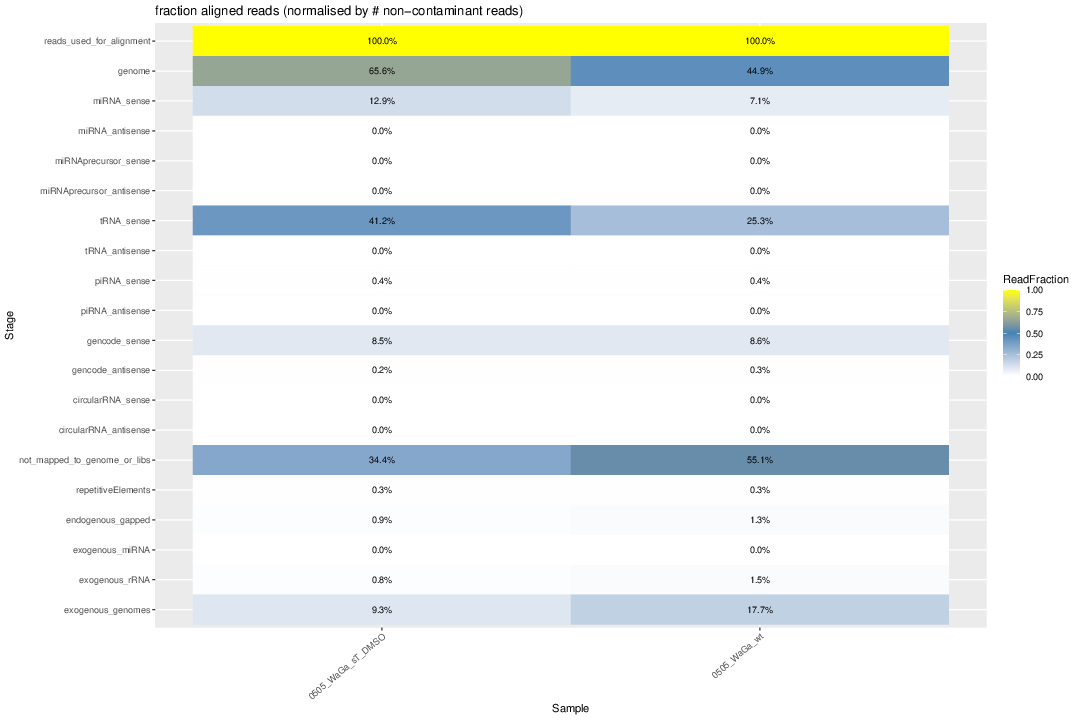
Re-draw the heatmap plots
#genome 97.9% 98.3% 21.3% 44.9% 81.4% 78.3% 78.5% 79.3% 73.3% 69.2% 65.6% 71.9% #miRNA_sense 84.7% 85.6% 3.5% 7.1% 16.2% 14.7% 15.8% 15.3% 7.5% 7.0% 12.9% 14.6% #miRNA_antisense 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% # #miRNAprecursor_sense 0.1% 0.1% 0.0% 0.0% 0.1% 0.1% 0.0% 0.1% 0.0% 0.0% 0.0% 0.0% #miRNAprecursor_antisense 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% # #tRNA_sense 3.4% 1.8% 8.4% 25.3% 45.3% 41.4% 48.8% 47.3% 52.1% 49.0% 41.2% 33.9% #tRNA_antisense 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% # #piRNA_sense 0.6% 0.5% 0.1% 0.4% 0.3% 0.4% 0.5% 0.4% 0.4% 0.5% 0.4% 0.6% #piRNA_antisense 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% # #gencode_sense 7.0% 8.5% 6.7% 8.6% 15.7% 16.6% 10.8% 12.9% 11.2% 10.8% 8.5% 18.3% #gencode_antisense 0.1% 0.1% 0.7% 0.3% 0.2% 0.3% 0.2% 0.2% 0.2% 0.2% 0.2% 0.3% #gencode 7.10% 8.60% 7.40% 8.90% 15.90% 16.90% 11.00% 13.10% 11.40% 11.00% 8.70% 18.60% # #circularRNA_sense 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% #circularRNA_antisense 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% 0.0% # #not_mapped_to_genome_or_libs 2.1% 1.7% 78.7% 55.1% 18.6% 21.7% 21.5% 20.7% 26.7% 30.8% 34.4% 28.1% import pandas as pd import numpy as np import seaborn as sns import matplotlib.pyplot as plt # Define data samples = [ "control MKL1", "control WaGa", "WaGa wildtype 0505", "WaGa wildtype 1905", "WaGa sT DMSO 0505", "WaGa sT DMSO 1905", "WaGa sT Dox 0505", "WaGa sT Dox 1905", "WaGa scr DMSO 0505", "WaGa scr DMSO 1905", "WaGa scr Dox 0505", "WaGa scr Dox 1905" ] #TODO_2: genome --> human_genome, not_mapped_to_genome_or_libs --> not_mapped_to_human_genome # send the new results including exogenous alignments to Ute! #categories = [ # "reads_used_for_alignment", "genome", "miRNA", "miRNAprecursor", "tRNA", "piRNA", # "gencode", "circularRNA", "not_mapped_to_genome_or_libs" #] categories = [ "reads_used_for_alignment", "human_genome", "miRNA", "miRNAprecursor", "tRNA", "piRNA", "gencode", "circularRNA", "not_mapped_to_human_genome" ] data = np.array([ [100.0, 100.0, 100.0, 100.0, 100.0, 100.0, 100.0, 100.0, 100.0, 100.0, 100.0, 100.0], [97.9, 98.3, 44.9, 21.3, 65.6, 71.9, 78.5, 81.4, 73.3, 79.3, 69.2, 78.3], [84.7, 85.6, 7.1, 3.5, 12.9, 14.6, 15.8, 16.2, 7.5, 15.3, 7.0, 14.7], [0.1, 0.1, 0.0, 0.0, 0.0, 0.0, 0.0, 0.1, 0.0, 0.1, 0.0, 0.1], [3.4, 1.8, 25.3, 8.4, 41.2, 33.9, 48.8, 45.3, 52.1, 47.3, 49.0, 41.4], [0.6, 0.5, 0.4, 0.1, 0.4, 0.6, 0.5, 0.3, 0.4, 0.4, 0.5, 0.4], [7.1, 8.6, 8.9, 7.4, 8.7, 18.6, 11.0, 15.9, 11.4, 13.1, 11.0, 16.9], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [2.1, 1.7, 55.1, 78.7, 34.4, 28.1, 21.5, 18.6, 26.7, 20.7, 30.8, 21.7] ]) ## Load data from Excel file #file_path = "mapping_heatmap.xlsx" # ## Read Excel file, assuming first column is index (row labels) #df = pd.read_excel(file_path, index_col=0) # Convert percentages to decimals data = data / 100.0 # Create DataFrame df = pd.DataFrame(data, index=categories, columns=samples) # Plot heatmap plt.figure(figsize=(14, 6)) sns.heatmap(df, annot=True, cmap="coolwarm", fmt=".3f", linewidths=0.5, cbar_kws={'label': 'Fraction Aligned Reads'}) # Improve layout plt.title("Heatmap of Read Alignments by Category and Sample", fontsize=14) plt.xlabel("Sample", fontsize=12) plt.ylabel("Read Category", fontsize=12) plt.xticks(rotation=15, ha="right", fontsize=10) plt.yticks(rotation=0, fontsize=10) plt.tight_layout() # Save as PNG plt.savefig("mapping_heatmap.png", dpi=300, bbox_inches="tight") # Show plot plt.show() -
Key steps of log: This log details the execution of a small RNA sequencing data analysis pipeline using the exceRpt tool (version 4.6.3) in a Docker container. The pipeline processes a human small RNA-seq dataset (testData_human.fastq.gz) with the following key steps:
-
Initial Setup
- Docker container launched with mounted volumes for input/output and reference databases.
- Parameters: hg38 genome, 50 threads, 200GB Java memory, exogenous mapping enabled.
- Docker container launched with input/output volume mounts
- 50 threads allocated with 200GB Java memory
- hg38 reference genome specified
-
Preprocessing
- Adapter detection and trimming using known adapter sequences.
- Quality filtering (Phred score ≥20, length ≥18nt).
- Removal of homopolymer-rich reads and low-quality sequences.
- Input FASTQ file decompressed (testData_human.fastq.gz)
- Adapter sequences identified using adapters.fa
- Quality encoding determined (Phred+33/64)
- Adapter clipping performed (TCGTATGCCGTCTTCTGCTTG)
- Quality filtering (Q20, p<80%)
- Homopolymer repeats filtered (max 66% single nt)
-
Contaminant Filtering
- Alignment against UniVec contaminants and ribosomal RNA (rRNA) databases.
- 322 reads processed, with statistics tracked at each step.
-
Endogenous RNA Analysis
- Alignment to human genome (hg38) and transcriptome.
- Quantification of small RNA types:
- miRNA (mature/precursor): Sense strands detected (antisense absent).
- tRNA, piRNA, gencode transcripts: Only sense strands reported.
- circRNA: Not detected in this dataset.
- Coverage and complexity metrics calculated.
-
Exogenous RNA Analysis
- Screened for microbial/viral RNAs:
- miRNA databases (miRBase).
- Ribosomal RNA databases.
- Comprehensive genomic databases (bacteria, plants, metazoa, fungi, viruses).
- Taxonomic classification of exogenous hits performed.
- Screened for microbial/viral RNAs:
-
QC & Results
- QC Result: PASS (based on transcriptome/genome ratio >0.5 and >100k transcriptome reads).
- Key Metrics:
- Input Reads: ~1.5 million (exact count not shown in log).
- Genome Mapped: Majority of reads.
- Transcriptome Complexity: Calculated ratio.
- Core results compressed into testData_human.fastq_CORE_RESULTS_v4.6.3.tgz.
-
Notable Observations:
- Antisense Reads: Absent for miRNA, tRNA, and piRNA (common in small RNA-seq).
- Potential Issues: Some files (e.g., antisense counts) were missing but did not disrupt pipeline.
- Resource Usage: High RAM (200GB) and multi-threading (50 cores) employed for efficiency.
-
Output Files:
- Quantified counts for endogenous RNAs (miRNA, tRNA, etc.).
- Exogenous RNA alignments with taxonomic annotations.
- QC report, adapter sequences, and alignment statistics.
-
-
Raw LOG of the pipeline providing a comprehensive small RNA profile, distinguishing host transcripts from contaminants and exogenous RNAs.
jhuang@WS-2290C:/media/jhuang/Elements/Data_Ute/Data_Ute_smallRNA_7$ docker run -v ~/DATA/Data_Ute/Data_Ute_smallRNA_7/results_exo4:/exceRptOutput -v /mnt/nvme0n1p1/MyexceRptDatabase:/exceRpt_DB -t rkitchen/excerpt INPUT_FILE_PATH=/exceRptInput/testData_human.fastq.gz MAIN_ORGANISM_GENOME_ID=hg38 N_THREADS=50 JAVA_RAM='200G' MAP_EXOGENOUS=on # mkdir -p /exceRptOutput/testData_human.fastq # gunzip -c /exceRptInput/testData_human.fastq.gz 2>> /exceRptOutput/testData_human.fastq.err | java -Xmx200G -jar /exceRpt_bin/exceRpt_Tools.jar FindAdapter -n 10000 -m 1000000 -s 4 -a /exceRpt_DB/adapters/adapters.fa - > /exceRptOutput/testData_human.fastq/testData_human.fastq.adapterSeq 2>> /exceRptOutput/testData_human.fastq.log # ## ASCII 84 is equal to Q20 (p<0.01) in Phred+64, so any file with max quals greater than this can reasonably assumed to be Phred+64 gunzip -c /exceRptInput/testData_human.fastq.gz | head -n 40000 | awk '{if(NR%4==0) printf("%s",$0);}' | od -A n -t u1 | grep -v "^\*" | awk 'BEGIN{min=100;max=0;}{for(i=1;i<=NF;i++) {if($i>max) max=$i; if($i<min) min=$i;}}END{if(max<84) print "33"; else print "64";}' > /exceRptOutput/testData_human.fastq/testData_human.fastq.qualityEncoding cat: /exceRptOutput/testData_human.fastq/testData_human.fastq.knownAdapterSeq: No such file or directory ## Run the SW alignment of known adapters regardless of user preference gunzip -c /exceRptInput/testData_human.fastq.gz 2>> /exceRptOutput/testData_human.fastq.err | java -Xmx200G -jar /exceRpt_bin/exceRpt_Tools.jar FindAdapter -n 1000 -m 100000 -s 4 -a /exceRpt_DB/adapters/adapters.fa - > /exceRptOutput/testData_human.fastq/testData_human.fastq.knownAdapterSeq 2>> /exceRptOutput/testData_human.fastq.log #@echo -e "`/bin/date "+%Y-%m-%d--%H:%M:%S"` exceRpt_smallRNA: Known adapter sequence: \n" >> /exceRptOutput/testData_human.fastq.log ## Carry on with the adapter provided / guessed gunzip -c /exceRptInput/testData_human.fastq.gz > /exceRptOutput/testData_human.fastq/testData_human.fastq.preClipped.fastq.tmp; /exceRpt_bin/fastx_0.0.14/bin/fastx_clipper -Q33 -a TCGTATGCCGTCTTCTGCTTG -l 18 -v -n -M 7 -i /exceRptOutput/testData_human.fastq/testData_human.fastq.preClipped.fastq.tmp -z -o /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.fastq.tmp.gz >> /exceRptOutput/testData_human.fastq.log 2>> /exceRptOutput/testData_human.fastq.err; rm /exceRptOutput/testData_human.fastq/testData_human.fastq.preClipped.fastq.tmp ## Count reads input to adapter clipping grep "Input: " /exceRptOutput/testData_human.fastq.log | awk '{print "input\t"$2}' >> /exceRptOutput/testData_human.fastq.stats ## Count reads output following adapter clipping grep "Output: " /exceRptOutput/testData_human.fastq.log | awk '{print "successfully_clipped\t"$2}' >> /exceRptOutput/testData_human.fastq.stats ## Remove random barcodes if there are any mv /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.fastq.tmp.gz /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.fastq.gz gunzip -c /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.fastq.gz | java -Xmx200G -jar /exceRpt_bin/exceRpt_Tools.jar TrimFastq -5p 0 -3p 0 | gzip -c > /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.fastq.gz 2>>/exceRptOutput/testData_human.fastq.log gunzip -c /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.fastq.gz | /exceRpt_bin/fastx_0.0.14/bin/fastq_quality_filter -v -Q33 -p 80 -q 20 > /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.tmp 2>>/exceRptOutput/testData_human.fastq.log ## Count reads that failed the quality filter grep "low-quality reads" /exceRptOutput/testData_human.fastq.log | awk '{print "failed_quality_filter\t"$2}' >> /exceRptOutput/testData_human.fastq.stats # # Filter homopolymer reads (those that have too many single nt repeats) java -Xmx200G -jar /exceRpt_bin/exceRpt_Tools.jar RemoveHomopolymerRepeats --verbose -m 0.66 -i /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.tmp -o /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.fastq >> /exceRptOutput/testData_human.fastq.log 2>> /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.REMOVEDRepeatReads.fastq gzip /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.REMOVEDRepeatReads.fastq gzip /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.fastq rm /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.tmp ## Count homopolymer repeat reads that failed the quality filter grep "Done. Sequences removed" /exceRptOutput/testData_human.fastq.log | awk -F "=" '{print "failed_homopolymer_filter\t"$2}' >> /exceRptOutput/testData_human.fastq.stats gunzip -c /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.fastq.gz > /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.fastq java -Xmx200G -jar /exceRpt_bin/exceRpt_Tools.jar GetSequenceLengths /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.fastq > /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.readLengths.txt 2>> /exceRptOutput/testData_human.fastq.err rm /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.fastq java -classpath /exceRpt_bin/FastQC_0.11.7:/exceRpt_bin/FastQC_0.11.7/sam-1.103.jar:/exceRpt_bin/FastQC_0.11.7/jbzip2-0.9.jar -Xmx200G -Dfastqc.threads=50 -Dfastqc.unzip=false -Dfastqc.output_dir=/exceRptOutput/testData_human.fastq/ uk/ac/babraham/FastQC/FastQCApplication /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.fastq.gz >> /exceRptOutput/testData_human.fastq.log 2>> /exceRptOutput/testData_human.fastq.err ## Count calibrator oligo reads echo -e "calibrator\tNA" >> /exceRptOutput/testData_human.fastq.stats # /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/filteringAlignments_UniVec_ --genomeDir /exceRpt_DB/UniVec/STAR_INDEX_UniVec --readFilesIn /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.fastq.gz --outReadsUnmapped Fastx --parametersFiles /exceRpt_DB/STAR_Parameters_Endogenous_smallRNA.in --alignEndsType Local --outFilterMatchNmin 18 --outFilterMatchNminOverLread 0.9 --outFilterMismatchNmax 1 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log 2>> /exceRptOutput/testData_human.fastq.err; /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/filteringAlignments_UniVec_Aligned.out.bam | awk '{print $3}' | sort -k 2,2 2>> /exceRptOutput/testData_human.fastq.err | uniq --count > /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.uniVecContaminants.counts 2>> /exceRptOutput/testData_human.fastq.err; /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/filteringAlignments_UniVec_Aligned.out.bam | awk '{print $1}' | sort 2>> /exceRptOutput/testData_human.fastq.err | uniq -c | wc -l > /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.uniVecContaminants.readCount 2>> /exceRptOutput/testData_human.fastq.err; gzip -c /exceRptOutput/testData_human.fastq/filteringAlignments_UniVec_Unmapped.out.mate1 > /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.noUniVecContaminants.fastq.gz; rm /exceRptOutput/testData_human.fastq/filteringAlignments_UniVec_Unmapped.out.mate1 ## Count UniVec contaminant reads cat /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.uniVecContaminants.readCount | awk '{print "UniVec_contaminants\t"$1}' >> /exceRptOutput/testData_human.fastq.stats /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/filteringAlignments_rRNA_ --genomeDir /exceRpt_DB/hg38/STAR_INDEX_rRNA --readFilesIn /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.noUniVecContaminants.fastq.gz --outReadsUnmapped Fastx --parametersFiles /exceRpt_DB/STAR_Parameters_Endogenous_smallRNA.in --alignEndsType Local --outFilterMatchNmin 18 --outFilterMatchNminOverLread 0.9 --outFilterMismatchNmax 1 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log 2>> /exceRptOutput/testData_human.fastq.err; /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/filteringAlignments_rRNA_Aligned.out.bam | awk '{print $3}' | sort -k 2,2 2>> /exceRptOutput/testData_human.fastq.err | uniq -c > /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.rRNA.counts 2>> /exceRptOutput/testData_human.fastq.err; /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/filteringAlignments_rRNA_Aligned.out.bam | awk '{print $1}' | sort 2>> /exceRptOutput/testData_human.fastq.err | uniq -c | wc -l > /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.rRNA.readCount 2>> /exceRptOutput/testData_human.fastq.err; gzip -c /exceRptOutput/testData_human.fastq/filteringAlignments_rRNA_Unmapped.out.mate1 > /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.noRiboRNA.fastq.gz; rm /exceRptOutput/testData_human.fastq/filteringAlignments_rRNA_Unmapped.out.mate1 ## Count rRNA reads cat /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.rRNA.readCount | awk ' {print "rRNA\t"$1}' >> /exceRptOutput/testData_human.fastq.stats # /exceRpt_bin/samtools-1.7/samtools sort -@ 50 -m 2G -O bam -T /exceRptOutput/testData_human.fastq/tmp /exceRptOutput/testData_human.fastq/filteringAlignments_rRNA_Aligned.out.bam > /exceRptOutput/testData_human.fastq/filteringAlignments_rRNA_Aligned.out.sorted.bam /exceRpt_bin/samtools-1.7/samtools index /exceRptOutput/testData_human.fastq/filteringAlignments_rRNA_Aligned.out.sorted.bam rm /exceRptOutput/testData_human.fastq/filteringAlignments_rRNA_Aligned.out.bam java -classpath /exceRpt_bin/FastQC_0.11.7:/exceRpt_bin/FastQC_0.11.7/sam-1.103.jar:/exceRpt_bin/FastQC_0.11.7/jbzip2-0.9.jar -Xmx200G -Dfastqc.threads=50 -Dfastqc.unzip=false -Dfastqc.output_dir=/exceRptOutput/testData_human.fastq/ uk/ac/babraham/FastQC/FastQCApplication /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.noRiboRNA.fastq.gz >> /exceRptOutput/testData_human.fastq.log 2>> /exceRptOutput/testData_human.fastq.err /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/endogenousAlignments_genome_ --genomeDir /exceRpt_DB/hg38/STAR_INDEX_genome --readFilesIn /exceRptOutput/testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.noRiboRNA.fastq.gz --outReadsUnmapped Fastx --parametersFiles /exceRpt_DB/STAR_Parameters_Endogenous_smallRNA.in --alignEndsType Local --outFilterMatchNmin 18 --outFilterMatchNminOverLread 0.9 --outFilterMismatchNmax 1 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log 2>> /exceRptOutput/testData_human.fastq.err # ## sort the alignments by ReadID just in case these are paired end reads in a single file? -- no, better to flag that this is an invalid file (ToDo) # ## v use this line when we start dealing with paired-end reads #/exceRpt_bin/samtools-1.7/samtools fastq -1 /exceRptOutput/testData_human.fastq/endogenousAlignments_genome_Mapped.out.mate1 -2 /exceRptOutput/testData_human.fastq/endogenousAlignments_genome_Mapped.out.mate2 /exceRptOutput/testData_human.fastq/endogenousAlignments_genome_Aligned.out.bam /exceRpt_bin/samtools-1.7/samtools fastq /exceRptOutput/testData_human.fastq/endogenousAlignments_genome_Aligned.out.bam > /exceRptOutput/testData_human.fastq/endogenousAlignments_genome_Mapped.out.mate1 [M::bam2fq_mainloop] discarded 0 singletons [M::bam2fq_mainloop] processed 322 reads # ## map ALL READS to the TRANSCRIPTOME (STAR ungapped) /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeMapped_transcriptome_ --readFilesIn /exceRptOutput/testData_human.fastq/endogenousAlignments_genome_Mapped.out.mate1 --genomeDir /exceRpt_DB/hg38/STAR_INDEX_transcriptome --parametersFiles /exceRpt_DB/STAR_Parameters_Endogenous_smallRNA.in --alignEndsType Local --outFilterMatchNmin 18 --outFilterMatchNminOverLread 0.9 --outFilterMismatchNmax 1 --outFilterMismatchNoverLmax 0.3 --readFilesCommand - >> /exceRptOutput/testData_human.fastq.log 2>> /exceRptOutput/testData_human.fastq.err gzip -c /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeMapped_transcriptome_Unmapped.out.mate1 > /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeMapped_transcriptome_Unmapped.R1.fastq.gz # /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeUnmapped_transcriptome_ --readFilesIn /exceRptOutput/testData_human.fastq/endogenousAlignments_genome_Unmapped.out.mate1 --outReadsUnmapped Fastx --genomeDir /exceRpt_DB/hg38/STAR_INDEX_transcriptome --parametersFiles /exceRpt_DB/STAR_Parameters_Endogenous_smallRNA.in --alignEndsType Local --outFilterMatchNmin 18 --outFilterMatchNminOverLread 0.9 --outFilterMismatchNmax 1 --outFilterMismatchNoverLmax 0.3 --readFilesCommand - >> /exceRptOutput/testData_human.fastq.log 2>> /exceRptOutput/testData_human.fastq.err gzip -c /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeUnmapped_transcriptome_Unmapped.out.mate1 > /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeUnmapped_transcriptome_Unmapped.R1.fastq.gz # ## Count # mapped reads cat /exceRptOutput/testData_human.fastq/endogenousAlignments_genome_Log.final.out | grep "Number of input reads" | awk -F "|\t" '{print "reads_used_for_alignment\t"$2}' >> /exceRptOutput/testData_human.fastq.stats cat /exceRptOutput/testData_human.fastq/endogenousAlignments_genome*apped_transcriptome_Log.final.out | grep "Number of input reads\|Uniquely mapped reads number\|Number of reads mapped to multiple loci" | sed '2,4d' | awk -F "|\t" '{SUM+=$2}END{print "genome\t"SUM}' >> /exceRptOutput/testData_human.fastq.stats # ## Compress STAR logs gzip /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeMapped_transcriptome_Log.out gzip /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeUnmapped_transcriptome_Log.out # ## Tidy up rm /exceRptOutput/testData_human.fastq/endogenousAlignments_genome_SJ.out.tab rm /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeUnmapped_transcriptome_SJ.out.tab rm /exceRptOutput/testData_human.fastq/endogenousAlignments_genome_Mapped.out.mate1 rm /exceRptOutput/testData_human.fastq/endogenousAlignments_genome_Unmapped.out.mate1 rm /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeMapped_transcriptome_Unmapped.out.mate1 rm /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeUnmapped_transcriptome_Unmapped.out.mate1 # java -Xmx200G -jar /exceRpt_bin/exceRpt_Tools.jar CIGAR_2_PWM -f /exceRptOutput/testData_human.fastq/endogenousAlignments_genome_Aligned.out.bam > /exceRptOutput/testData_human.fastq/endogenousAlignments_genome_Aligned.out.bam.CIGARstats.txt 2>> /exceRptOutput/testData_human.fastq.log # /exceRpt_bin/samtools-1.7/samtools sort -n -@ 50 -m 2G -O bam -T /exceRptOutput/testData_human.fastq/tmp /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeMapped_transcriptome_Aligned.out.bam > /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeMapped_transcriptome_Aligned.out.sorted.bam 2>> /exceRptOutput/testData_human.fastq.log # java -Xmx200G -jar /exceRpt_bin/exceRpt_Tools.jar ReadCoverage -exceRpt -a /exceRpt_DB/hg38/gencodeAnnotation.gtf -f /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeMapped_transcriptome_Aligned.out.sorted.bam 2>> /exceRptOutput/testData_human.fastq.log # rm /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeMapped_transcriptome_Aligned.out.sorted.bam # ## Assign reads java -Xmx200G -jar /exceRpt_bin/exceRpt_Tools.jar ProcessEndogenousAlignments --libPriority miRNA,tRNA,piRNA,gencode,circRNA --genomeMappedReads /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeMapped_transcriptome_Aligned.out.bam --transcriptomeMappedReads /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeUnmapped_transcriptome_Aligned.out.bam --hairpin2genome /exceRpt_DB/hg38/miRNA_precursor2genome.sam --mature2hairpin /exceRpt_DB/hg38/miRNA_mature2precursor.sam --dict /exceRptOutput/testData_human.fastq/endogenousAlignments_Accepted.dict 2>> /exceRptOutput/testData_human.fastq.log | sort -k 2,2 -k 1,1 > /exceRptOutput/testData_human.fastq/endogenousAlignments_Accepted.txt # ## Do we want to downsample? # ## Quantify all annotated RNAs java -Xmx200G -jar /exceRpt_bin/exceRpt_Tools.jar QuantifyEndogenousAlignments --dict /exceRptOutput/testData_human.fastq/endogenousAlignments_Accepted.dict --acceptedAlignments /exceRptOutput/testData_human.fastq/endogenousAlignments_Accepted.txt --outputPath /exceRptOutput/testData_human.fastq 2>> /exceRptOutput/testData_human.fastq.log # ## Summarise alignment statistics cat /exceRptOutput/testData_human.fastq/readCounts_miRNAmature_sense.txt | awk '{SUM+=$4}END{printf "miRNA_sense\t%.0f\n",SUM}' >> /exceRptOutput/testData_human.fastq.stats cat /exceRptOutput/testData_human.fastq/readCounts_miRNAmature_antisense.txt | awk '{SUM+=$4}END{printf "miRNA_antisense\t%.0f\n",SUM}' >> /exceRptOutput/testData_human.fastq.stats cat: /exceRptOutput/testData_human.fastq/readCounts_miRNAmature_antisense.txt: No such file or directory cat /exceRptOutput/testData_human.fastq/readCounts_miRNAprecursor_sense.txt | awk '{SUM+=$4}END{printf "miRNAprecursor_sense\t%.0f\n",SUM}' >> /exceRptOutput/testData_human.fastq.stats cat /exceRptOutput/testData_human.fastq/readCounts_miRNAprecursor_antisense.txt | awk '{SUM+=$4}END{printf "miRNAprecursor_antisense\t%.0f\n",SUM}' >> /exceRptOutput/testData_human.fastq.stats cat: /exceRptOutput/testData_human.fastq/readCounts_miRNAprecursor_antisense.txt: No such file or directory cat /exceRptOutput/testData_human.fastq/readCounts_tRNA_sense.txt | awk '{SUM+=$4}END{printf "tRNA_sense\t%.0f\n",SUM}' >> /exceRptOutput/testData_human.fastq.stats cat /exceRptOutput/testData_human.fastq/readCounts_tRNA_antisense.txt | awk '{SUM+=$4}END{printf "tRNA_antisense\t%.0f\n",SUM}' >> /exceRptOutput/testData_human.fastq.stats cat: /exceRptOutput/testData_human.fastq/readCounts_tRNA_antisense.txt: No such file or directory cat /exceRptOutput/testData_human.fastq/readCounts_piRNA_sense.txt | awk '{SUM+=$4}END{printf "piRNA_sense\t%.0f\n",SUM}' >> /exceRptOutput/testData_human.fastq.stats cat /exceRptOutput/testData_human.fastq/readCounts_piRNA_antisense.txt | awk '{SUM+=$4}END{printf "piRNA_antisense\t%.0f\n",SUM}' >> /exceRptOutput/testData_human.fastq.stats cat: /exceRptOutput/testData_human.fastq/readCounts_piRNA_antisense.txt: No such file or directory cat /exceRptOutput/testData_human.fastq/readCounts_gencode_sense.txt | awk '{SUM+=$4}END{printf "gencode_sense\t%.0f\n",SUM}' >> /exceRptOutput/testData_human.fastq.stats cat /exceRptOutput/testData_human.fastq/readCounts_gencode_antisense.txt | awk '{SUM+=$4}END{printf "gencode_antisense\t%.0f\n",SUM}' >> /exceRptOutput/testData_human.fastq.stats cat /exceRptOutput/testData_human.fastq/readCounts_circRNA_sense.txt | awk '{SUM+=$4}END{printf "circularRNA_sense\t%.0f\n",SUM}' >> /exceRptOutput/testData_human.fastq.stats cat: /exceRptOutput/testData_human.fastq/readCounts_circRNA_sense.txt: No such file or directory cat /exceRptOutput/testData_human.fastq/readCounts_circRNA_antisense.txt | awk '{SUM+=$4}END{printf "circularRNA_antisense\t%.0f\n",SUM}' >> /exceRptOutput/testData_human.fastq.stats cat: /exceRptOutput/testData_human.fastq/readCounts_circRNA_antisense.txt: No such file or directory ## Count reads not mapping to the genome or to the libraries gunzip -c /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeUnmapped_transcriptome_Unmapped.R1.fastq.gz | wc -l | awk '{print "not_mapped_to_genome_or_libs\t"($1/4)}' >> /exceRptOutput/testData_human.fastq.stats # ## Tidy up gzip -c /exceRptOutput/testData_human.fastq/endogenousAlignments_Accepted.txt > /exceRptOutput/testData_human.fastq/endogenousAlignments_Accepted.txt.gz rm /exceRptOutput/testData_human.fastq/endogenousAlignments_Accepted.txt /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/endogenousAlignments_repetitiveElements_ --genomeDir /exceRpt_DB/hg38/STAR_INDEX_repetitiveElements --readFilesIn /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeUnmapped_transcriptome_Unmapped.R1.fastq.gz --outReadsUnmapped Fastx --parametersFiles /exceRpt_DB/STAR_Parameters_Endogenous_smallRNA.in --alignEndsType Local --outFilterMatchNmin 18 --outFilterMatchNminOverLread 0.9 --outFilterMismatchNmax 1 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log 2>> /exceRptOutput/testData_human.fastq.err ## Assigned non-redundantly to annotated REs /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/endogenousAlignments_repetitiveElements_Aligned.out.bam | grep -v "^@" | awk '{print $1}' | sort | uniq | wc -l | awk '{print "repetitiveElements\t"$0}' >> /exceRptOutput/testData_human.fastq.stats gzip -c /exceRptOutput/testData_human.fastq/endogenousAlignments_repetitiveElements_Unmapped.out.mate1 > /exceRptOutput/testData_human.fastq/endogenousAlignments_repetitiveElements_Unmapped.R1.fastq.gz /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeGapped_ --alignIntronMax 0 --alignIntronMin 21 --genomeDir /exceRpt_DB/hg38/STAR_INDEX_genome --readFilesIn /exceRptOutput/testData_human.fastq/endogenousAlignments_repetitiveElements_Unmapped.R1.fastq.gz --outReadsUnmapped Fastx --parametersFiles /exceRpt_DB/STAR_Parameters_Endogenous_smallRNA.in --alignEndsType Local --outFilterMatchNmin 18 --outFilterMatchNminOverLread 0.9 --outFilterMismatchNmax 1 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log 2>> /exceRptOutput/testData_human.fastq.err ## mapped to the genome with gaps /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeGapped_Aligned.out.bam | grep -v "^@" | awk '{print $1}' | sort | uniq | wc -l | awk '{print "endogenous_gapped\t"$0}' >> /exceRptOutput/testData_human.fastq.stats gzip -c /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeGapped_Unmapped.out.mate1 > /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeGapped_Unmapped.R1.fastq.gz mkdir -p /exceRptOutput/testData_human.fastq/EXOGENOUS_miRNA /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_miRNA/exogenous_miRBase_ --genomeDir /exceRpt_DB/miRBase/STAR_INDEX_miRBaseAll --readFilesIn /exceRptOutput/testData_human.fastq/endogenousAlignments_genomeGapped_Unmapped.R1.fastq.gz --outReadsUnmapped Fastx --parametersFiles /exceRpt_DB/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log 2>> /exceRptOutput/testData_human.fastq.err gzip -c /exceRptOutput/testData_human.fastq/EXOGENOUS_miRNA/exogenous_miRBase_Unmapped.out.mate1 > /exceRptOutput/testData_human.fastq/EXOGENOUS_miRNA/exogenous_miRBase_Unmapped.R1.fastq.gz rm /exceRptOutput/testData_human.fastq/EXOGENOUS_miRNA/exogenous_miRBase_Unmapped.out.mate1 # ## quantify read alignments using a slight hack of the endogenous alignment engine java -Xmx200G -jar /exceRpt_bin/exceRpt_Tools.jar ProcessEndogenousAlignments --forceLib miRNA --transcriptomeMappedReads /exceRptOutput/testData_human.fastq/EXOGENOUS_miRNA/exogenous_miRBase_Aligned.out.bam --hairpin2genome /exceRpt_DB/miRBase/miRNA_precursor2genome.sam --mature2hairpin /exceRpt_DB/miRBase/miRNA_mature2precursor.sam --dict /exceRptOutput/testData_human.fastq/EXOGENOUS_miRNA/exogenousAlignments_Accepted.dict 2>> /exceRptOutput/testData_human.fastq.log | sort -k 2,2 -k 1,1 > /exceRptOutput/testData_human.fastq/EXOGENOUS_miRNA/exogenousAlignments_Accepted.txt # java -Xmx200G -jar /exceRpt_bin/exceRpt_Tools.jar QuantifyEndogenousAlignments --dict /exceRptOutput/testData_human.fastq/EXOGENOUS_miRNA/exogenousAlignments_Accepted.dict --acceptedAlignments /exceRptOutput/testData_human.fastq/EXOGENOUS_miRNA/exogenousAlignments_Accepted.txt --outputPath /exceRptOutput/testData_human.fastq/EXOGENOUS_miRNA 2>> /exceRptOutput/testData_human.fastq.log # ## Tidy up: gzip -c /exceRptOutput/testData_human.fastq/EXOGENOUS_miRNA/exogenousAlignments_Accepted.txt > /exceRptOutput/testData_human.fastq/EXOGENOUS_miRNA/exogenousMiRNAAlignments_Accepted.txt.gz rm /exceRptOutput/testData_human.fastq/EXOGENOUS_miRNA/exogenousAlignments_Accepted.txt # ## Stats cat /exceRptOutput/testData_human.fastq/EXOGENOUS_miRNA/exogenous_miRBase_Log.final.out | grep "Number of input reads" | awk -F "|\t" '{print "input_to_exogenous_miRNA\t"$2}' >> /exceRptOutput/testData_human.fastq.stats cat /exceRptOutput/testData_human.fastq/EXOGENOUS_miRNA/exogenous_miRBase_Log.final.out | grep "Uniquely mapped reads number\|Number of reads mapped to multiple loci" | awk -F "|\t" '{SUM+=$2}END{print "exogenous_miRNA\t"SUM}' >> /exceRptOutput/testData_human.fastq.stats mkdir -p /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/exogenous_rRNA_ --genomeDir /exceRpt_DB/ribosomeDatabase/exogenous_rRNAs --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_miRNA/exogenous_miRBase_Unmapped.R1.fastq.gz --outReadsUnmapped Fastx --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log ## Input to exogenous rRNA alignment grep "Number of input reads" /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/exogenous_rRNA_Log.final.out | tr '[:blank:]' ' ' | awk -F " \\\| " '{print "input_to_exogenous_rRNA\t"$2}' >> /exceRptOutput/testData_human.fastq.stats ## Assigned non-redundantly to annotated exogenous rRNAs cat /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/exogenous_rRNA_Log.final.out | grep "Uniquely mapped reads number\|Number of reads mapped to multiple loci" | awk -F "|\t" '{SUM+=$2}END{print "exogenous_rRNA\t"SUM}' >> /exceRptOutput/testData_human.fastq.stats #/exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/exogenous_rRNA_Aligned.out.bam | awk '{print $1}' | sort | uniq | wc -l | awk '{print "exogenous_rRNA\t"$0}' >> /exceRptOutput/testData_human.fastq.stats ## compress and tidy up gzip -c /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/exogenous_rRNA_Unmapped.out.mate1 > /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz rm /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/exogenous_rRNA_Unmapped.out.mate1 rm /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/exogenous_rRNA_Log.out /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/exogenous_rRNA_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | sort -k 1,1 -k 2,2 | uniq > /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/ExogenousRibosomalAlignments.txt 2>> /exceRptOutput/testData_human.fastq.log # java -Xmx200G -jar /exceRpt_bin/exceRpt_Tools.jar ProcessExogenousAlignments -taxonomyPath /exceRpt_DB/NCBI_taxonomy_taxdump -min 0.001 -frac 0.95 --minReads 3 -batchSize 20000 -alignments /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/ExogenousRibosomalAlignments.txt --rdp > /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/ExogenousRibosomalAlignments.tmp 2>> /exceRptOutput/testData_human.fastq.log # # Tidy up mv /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/ExogenousRibosomalAlignments.tmp /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/ExogenousRibosomalAlignments.result.taxaAnnotated.txt rm /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/ExogenousRibosomalAlignments.txt mkdir -p /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Bacteria1_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_BACTERIA1 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Bacteria2_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_BACTERIA2 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Bacteria3_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_BACTERIA3 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Bacteria4_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_BACTERIA4 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Bacteria5_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_BACTERIA5 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Bacteria6_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_BACTERIA6 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Bacteria7_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_BACTERIA7 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Bacteria8_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_BACTERIA8 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Bacteria9_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_BACTERIA9 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Bacteria10_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_BACTERIA10 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log mkdir -p /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Plants1_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_PLANTS1 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Plants2_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_PLANTS2 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Plants3_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_PLANTS3 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Plants4_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_PLANTS4 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Plants5_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_PLANTS5 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log mkdir -p /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Metazoa1_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_METAZOA1 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Metazoa2_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_METAZOA2 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Metazoa3_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_METAZOA3 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Metazoa4_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_METAZOA4 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Metazoa5_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_METAZOA5 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log mkdir -p /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/FungiProtistVirus_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_FUNGI_PROTIST_VIRUS --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log mkdir -p /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Vertebrate1_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_VERTEBRATE1 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Vertebrate2_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_VERTEBRATE2 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Vertebrate3_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_VERTEBRATE3 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log /exceRpt_bin/STAR-2.5.4b/bin/Linux_x86_64/STAR --runThreadN 50 --outFileNamePrefix /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Vertebrate4_ --genomeDir /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_GENOME_VERTEBRATE4 --readFilesIn /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz --parametersFiles /exceRpt_DB/Genomes_BacteriaFungiMammalPlantProtistVirus/STAR_Parameters_Exogenous.in --outSAMtype BAM Unsorted --outSAMattributes Standard --alignEndsType EndToEnd --outFilterMatchNmin 18 --outFilterMatchNminOverLread 1.0 --outFilterMismatchNmax 0 --outFilterMismatchNoverLmax 0.3 >> /exceRptOutput/testData_human.fastq.log # /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Bacteria1_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,"[$:]");print $1"\tBacteria\t"a[1]"\t"a[7]"\t"$3"\t"$4"\t"$5}' > /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Bacteria2_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,"[$:]");print $1"\tBacteria\t"a[1]"\t"a[7]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Bacteria3_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,"[$:]");print $1"\tBacteria\t"a[1]"\t"a[7]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Bacteria4_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,"[$:]");print $1"\tBacteria\t"a[1]"\t"a[7]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Bacteria5_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,"[$:]");print $1"\tBacteria\t"a[1]"\t"a[7]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Bacteria6_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,"[$:]");print $1"\tBacteria\t"a[1]"\t"a[7]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Bacteria7_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,"[$:]");print $1"\tBacteria\t"a[1]"\t"a[7]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Bacteria8_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,"[$:]");print $1"\tBacteria\t"a[1]"\t"a[7]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Bacteria9_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,"[$:]");print $1"\tBacteria\t"a[1]"\t"a[7]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Bacteria10_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,"[$:]");print $1"\tBacteria\t"a[1]"\t"a[7]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt # /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Plants1_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,":");print $1"\t"a[1]"\t"a[2]"\t"a[3]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Plants2_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,":");print $1"\t"a[1]"\t"a[2]"\t"a[3]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Plants3_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,":");print $1"\t"a[1]"\t"a[2]"\t"a[3]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Plants4_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,":");print $1"\t"a[1]"\t"a[2]"\t"a[3]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Plants5_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,":");print $1"\t"a[1]"\t"a[2]"\t"a[3]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt # /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Metazoa1_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,":");print $1"\t"a[1]"\t"a[2]"\t"a[3]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Metazoa2_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,":");print $1"\t"a[1]"\t"a[2]"\t"a[3]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Metazoa3_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,":");print $1"\t"a[1]"\t"a[2]"\t"a[3]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Metazoa4_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,":");print $1"\t"a[1]"\t"a[2]"\t"a[3]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Metazoa5_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,":");print $1"\t"a[1]"\t"a[2]"\t"a[3]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt # /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/FungiProtistVirus_Aligned.out.bam | grep "Virus:" | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,"[:|]");print $1"\t"a[1]"\t"a[3]"\t"a[5]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/FungiProtistVirus_Aligned.out.bam | grep "Fungi:" | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,":");print $1"\t"a[1]"\t"a[2]"\t"a[3]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/FungiProtistVirus_Aligned.out.bam | grep "Protist:" | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,":");print $1"\t"a[1]"\t"a[2]"\t"a[3]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt # /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Vertebrate1_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,":");print $1"\t"a[1]"\t"a[2]"\t"a[3]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Vertebrate2_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,":");print $1"\t"a[1]"\t"a[2]"\t"a[3]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Vertebrate3_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,":");print $1"\t"a[1]"\t"a[2]"\t"a[3]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt /exceRpt_bin/samtools-1.7/samtools view /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/Vertebrate4_Aligned.out.bam | awk '{print $1,$3,$4,$6,$10}' | uniq | awk '{split($2,a,":");print $1"\t"a[1]"\t"a[2]"\t"a[3]"\t"$3"\t"$4"\t"$5}' >> /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt # cat /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt | sort -k 1,1 | gzip -c > /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.sorted.txt.gz rm /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.txt # ## Input to exogenous genome alignment gunzip -c /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA/unaligned.fq.gz | wc -l | awk '{print "input_to_exogenous_genomes\t"$1/4}' >> /exceRptOutput/testData_human.fastq.stats ## Count reads mapped to exogenous genomes: gunzip -c /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.sorted.txt.gz | awk '{print $1}' | uniq | wc -l | awk '{print "exogenous_genomes\t"$1}' >> /exceRptOutput/testData_human.fastq.stats # gunzip -c /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.sorted.txt.gz > /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.sorted.txt java -Xmx200G -jar /exceRpt_bin/exceRpt_Tools.jar ProcessExogenousAlignments -taxonomyPath /exceRpt_DB/NCBI_taxonomy_taxdump -min 0.001 -frac 0.95 -batchSize 500000 -minReads 3 -alignments /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.sorted.txt > /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.tmp 2>> /exceRptOutput/testData_human.fastq.log # Tidy up mv /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.tmp /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.result.taxaAnnotated.txt rm /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.sorted.txt ## Wrap up logging and stats files # ## Adapter confidence echo -e "known: " >> /exceRptOutput/testData_human.fastq.qctmp cat /exceRptOutput/testData_human.fastq/testData_human.fastq.knownAdapterSeq >> /exceRptOutput/testData_human.fastq.qctmp echo -e "used: " >> /exceRptOutput/testData_human.fastq.qctmp cat /exceRptOutput/testData_human.fastq/testData_human.fastq.adapterSeq >> /exceRptOutput/testData_human.fastq.qctmp cat /exceRptOutput/testData_human.fastq.qctmp | tr '\n' ' ' | awk -F ' ' '{if($2=="used:"){ if(NF==2){print "Adapter_confidence: LOW"}else{print "Adapter_confidence: WARN_unableToGuessAdapter_usingProvided("$3")"}}else{if($2==$4){print "Adapter_confidence: HIGH"}else{print "Adapter_confidence: WARN_providedAdapter("$4")DisagreesWithGuessed("$2")"}}}' > /exceRptOutput/testData_human.fastq.qcResult # ## Calculate QC result cat /exceRptOutput/testData_human.fastq.stats | grep "^input" | head -n 1 | awk '{print $2}' > /exceRptOutput/testData_human.fastq.qctmp cat /exceRptOutput/testData_human.fastq.stats | grep "^genome" | awk '{print $2}' >> /exceRptOutput/testData_human.fastq.qctmp cat /exceRptOutput/testData_human.fastq.stats | grep "sense" | awk '{SUM+=$2}END{print SUM}' >> /exceRptOutput/testData_human.fastq.qctmp cat /exceRptOutput/testData_human.fastq.qctmp | tr '\n' '\t' | awk '{result="FAIL"; ratio=0; if($2>0){ratio=$3/$2}; if(ratio>0.5 && $3>100000)result="PASS"}END{print "QC_result: "result"\nInputReads: "$1"\nGenomeReads: "$2"\nTranscriptomeReads: "$3"\nTranscriptomeGenomeRatio: "ratio}' >> /exceRptOutput/testData_human.fastq.qcResult gunzip -c /exceRptOutput/testData_human.fastq/endogenousAlignments_Accepted.txt.gz | wc -l > /exceRptOutput/testData_human.fastq.qctmp gunzip -c /exceRptOutput/testData_human.fastq/endogenousAlignments_Accepted.txt.gz | awk '{print $2}' | uniq | wc -l >> /exceRptOutput/testData_human.fastq.qctmp # cat /exceRptOutput/testData_human.fastq.qctmp | tr '\n' '\t' | awk '{if($1>0){print "TranscriptomeComplexity: "($2/$1)}else{print "TranscriptomeComplexity: 0"}}' >> /exceRptOutput/testData_human.fastq.qcResult rm /exceRptOutput/testData_human.fastq.qctmp # ## Compress core results files automatically ls -lh /exceRptOutput/testData_human.fastq | awk '{print $9}' | grep "readCounts_\|.readLengths.txt\|_fastqc.zip\|.counts\|.knownAdapterSeq\|.adapterSeq\|.qualityEncoding\|.CIGARstats.txt\|.coverage.txt" | awk '{print "testData_human.fastq/"$1}' > /exceRptOutput/testData_human.fastq_filesToCompress.txt; echo testData_human.fastq.log >> /exceRptOutput/testData_human.fastq_filesToCompress.txt; echo testData_human.fastq.stats >> /exceRptOutput/testData_human.fastq_filesToCompress.txt; echo testData_human.fastq.qcResult >> /exceRptOutput/testData_human.fastq_filesToCompress.txt; ls -lh /exceRptOutput/testData_human.fastq | awk '{print $9}' | grep "calibratormapped.counts" | awk '{print "testData_human.fastq/"$1}' >> /exceRptOutput/testData_human.fastq_filesToCompress.txt; ls -lh /exceRptOutput/testData_human.fastq/EXOGENOUS_miRNA | awk '{print $9}' | grep "readCounts_" | awk '{print "testData_human.fastq/EXOGENOUS_miRNA/"$1}' >> /exceRptOutput/testData_human.fastq_filesToCompress.txt; ls -lh /exceRptOutput/testData_human.fastq/EXOGENOUS_rRNA | awk '{print $9}' | grep "ExogenousRibosomalAlignments.result.taxaAnnotated.txt" | awk '{print "testData_human.fastq/EXOGENOUS_rRNA/"$1}' >> /exceRptOutput/testData_human.fastq_filesToCompress.txt; ls -lh /exceRptOutput/testData_human.fastq/EXOGENOUS_genomes | awk '{print $9}' | grep "ExogenousGenomicAlignments.result.taxaAnnotated.txt" | awk '{print "testData_human.fastq/EXOGENOUS_genomes/"$1}' >> /exceRptOutput/testData_human.fastq_filesToCompress.txt tar -cvz -C /exceRptOutput -T /exceRptOutput/testData_human.fastq_filesToCompress.txt -f /exceRptOutput/testData_human.fastq_CORE_RESULTS_v4.6.3.tgz 2> /dev/null testData_human.fastq/endogenousAlignments_genomeMapped_transcriptome_Aligned.out.sorted.bam.coverage.txt testData_human.fastq/endogenousAlignments_genome_Aligned.out.bam.CIGARstats.txt testData_human.fastq/readCounts_gencode_antisense.txt testData_human.fastq/readCounts_gencode_antisense_geneLevel.txt testData_human.fastq/readCounts_gencode_sense.txt testData_human.fastq/readCounts_gencode_sense_geneLevel.txt testData_human.fastq/readCounts_miRNAmature_sense.txt testData_human.fastq/readCounts_miRNAprecursor_sense.txt testData_human.fastq/readCounts_piRNA_sense.txt testData_human.fastq/readCounts_tRNA_sense.txt testData_human.fastq/testData_human.fastq.adapterSeq testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.noRiboRNA_fastqc.zip testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.rRNA.counts testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.readLengths.txt testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered.uniVecContaminants.counts testData_human.fastq/testData_human.fastq.clipped.trimmed.filtered_fastqc.zip testData_human.fastq/testData_human.fastq.knownAdapterSeq testData_human.fastq/testData_human.fastq.qualityEncoding testData_human.fastq.log testData_human.fastq.stats testData_human.fastq.qcResult testData_human.fastq/EXOGENOUS_rRNA/ExogenousRibosomalAlignments.result.taxaAnnotated.txt testData_human.fastq/EXOGENOUS_genomes/ExogenousGenomicAlignments.result.taxaAnnotated.txt rm /exceRptOutput/testData_human.fastq_filesToCompress.txt ## END PIPELINE
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Kraken2 Installation and Usage Guide
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Should the inputs for GSVA be normalized or raw?
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- Risks of Rebooting into Rescue Mode
- 足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Processing Data_Michelle_RNAseq_2025
Setup the environment for lumicks-pylake and C_Trap-Multimer-photontrack.ipynb
🧬 Cadmium Resistance Gene Analysis in Staphylococcus epidermidis HD46
Workflow for RNA-Binding Protein Enrichment and RNA Type Distribution Analysis (Ute’s Project)