Processing Data_Tam_RNAseq_2025_LB_vs_Mac_ATCC19606
gene_x 0 like s 785 view s
Tags: pipeline, RNA-seq
-
Targets
Could you please assist me with processing RNA-seq data? The reference genome is CP059040. I aim to analyze the data using PCA, a Venn diagram, and KEGG and GO annotation enrichment analysis. The samples are labeled as follows (where 'x' indicates the replicate number): LB-AB-x LB-IJ-x LB-W1-x LB-WT19606-x LB-Y1-x Mac-AB-x Mac-IJ-x Mac-W1-x Mac-WT19606-x Mac-Y1-x -
Download the raw data
./lnd login -u X101SC25015922-Z02-J002 -p m*********5 ./lnd list ./lnd cp -d oss:// ./ ./lnd cp oss://CP2024102300053 . #Error ./lnd list oss://CP2024102300053 ./lnd cp -d oss://CP2024102300053/H101SC25015922/RSMR00204 . #CP2024102300053/H101SC25015922/RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002 -
Prepare raw data
mkdir raw_data; cd raw_data ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-AB-1/LB-AB-1_1.fq.gz LB-AB-r1_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-AB-1/LB-AB-1_2.fq.gz LB-AB-r1_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-AB-2/LB-AB-2_1.fq.gz LB-AB-r2_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-AB-2/LB-AB-2_2.fq.gz LB-AB-r2_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-AB-3/LB-AB-3_1.fq.gz LB-AB-r3_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-AB-3/LB-AB-3_2.fq.gz LB-AB-r3_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-IJ-1/LB-IJ-1_1.fq.gz LB-IJ-r1_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-IJ-1/LB-IJ-1_2.fq.gz LB-IJ-r1_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-IJ-2/LB-IJ-2_1.fq.gz LB-IJ-r2_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-IJ-2/LB-IJ-2_2.fq.gz LB-IJ-r2_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-IJ-4/LB-IJ-4_1.fq.gz LB-IJ-r4_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-IJ-4/LB-IJ-4_2.fq.gz LB-IJ-r4_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-W1-1/LB-W1-1_1.fq.gz LB-W1-r1_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-W1-1/LB-W1-1_2.fq.gz LB-W1-r1_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-W1-2/LB-W1-2_1.fq.gz LB-W1-r2_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-W1-2/LB-W1-2_2.fq.gz LB-W1-r2_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-W1-3/LB-W1-3_1.fq.gz LB-W1-r3_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-W1-3/LB-W1-3_2.fq.gz LB-W1-r3_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-WT19606-2/LB-WT19606-2_1.fq.gz LB-WT19606-r2_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-WT19606-2/LB-WT19606-2_2.fq.gz LB-WT19606-r2_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-WT19606-3/LB-WT19606-3_1.fq.gz LB-WT19606-r3_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-WT19606-3/LB-WT19606-3_2.fq.gz LB-WT19606-r3_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-WT19606-4/LB-WT19606-4_1.fq.gz LB-WT19606-r4_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-WT19606-4/LB-WT19606-4_2.fq.gz LB-WT19606-r4_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-Y1-2/LB-Y1-2_1.fq.gz LB-Y1-r2_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-Y1-2/LB-Y1-2_2.fq.gz LB-Y1-r2_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-Y1-3/LB-Y1-3_1.fq.gz LB-Y1-r3_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-Y1-3/LB-Y1-3_2.fq.gz LB-Y1-r3_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-Y1-4/LB-Y1-4_1.fq.gz LB-Y1-r4_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/LB-Y1-4/LB-Y1-4_2.fq.gz LB-Y1-r4_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-AB-1/Mac-AB-1_1.fq.gz Mac-AB-r1_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-AB-1/Mac-AB-1_2.fq.gz Mac-AB-r1_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-AB-2/Mac-AB-2_1.fq.gz Mac-AB-r2_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-AB-2/Mac-AB-2_2.fq.gz Mac-AB-r2_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-AB-3/Mac-AB-3_1.fq.gz Mac-AB-r3_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-AB-3/Mac-AB-3_2.fq.gz Mac-AB-r3_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-IJ-1/Mac-IJ-1_1.fq.gz Mac-IJ-r1_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-IJ-1/Mac-IJ-1_2.fq.gz Mac-IJ-r1_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-IJ-2/Mac-IJ-2_1.fq.gz Mac-IJ-r2_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-IJ-2/Mac-IJ-2_2.fq.gz Mac-IJ-r2_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-IJ-4/Mac-IJ-4_1.fq.gz Mac-IJ-r4_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-IJ-4/Mac-IJ-4_2.fq.gz Mac-IJ-r4_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-W1-1/Mac-W1-1_1.fq.gz Mac-W1-r1_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-W1-1/Mac-W1-1_2.fq.gz Mac-W1-r1_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-W1-2/Mac-W1-2_1.fq.gz Mac-W1-r2_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-W1-2/Mac-W1-2_2.fq.gz Mac-W1-r2_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-W1-3/Mac-W1-3_1.fq.gz Mac-W1-r3_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-W1-3/Mac-W1-3_2.fq.gz Mac-W1-r3_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-WT19606-2/Mac-WT19606-2_1.fq.gz Mac-WT19606-r2_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-WT19606-2/Mac-WT19606-2_2.fq.gz Mac-WT19606-r2_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-WT19606-3/Mac-WT19606-3_1.fq.gz Mac-WT19606-r3_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-WT19606-3/Mac-WT19606-3_2.fq.gz Mac-WT19606-r3_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-WT19606-4/Mac-WT19606-4_1.fq.gz Mac-WT19606-r4_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-WT19606-4/Mac-WT19606-4_2.fq.gz Mac-WT19606-r4_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-Y1-2/Mac-Y1-2_1.fq.gz Mac-Y1-r2_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-Y1-2/Mac-Y1-2_2.fq.gz Mac-Y1-r2_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-Y1-3/Mac-Y1-3_1.fq.gz Mac-Y1-r3_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-Y1-3/Mac-Y1-3_2.fq.gz Mac-Y1-r3_R2.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-Y1-4/Mac-Y1-4_1.fq.gz Mac-Y1-r4_R1.fq.gz ln -s ../RSMR00204/X101SC25015922-Z02/X101SC25015922-Z02-J002/01.RawData/Mac-Y1-4/Mac-Y1-4_2.fq.gz Mac-Y1-r4_R2.fq.gz -
Preparing the directory trimmed
mkdir trimmed trimmed_unpaired; for sample_id in LB-AB-r1 LB-AB-r2 LB-AB-r3 LB-IJ-r1 LB-IJ-r2 LB-IJ-r4 LB-W1-r1 LB-W1-r2 LB-W1-r3 LB-WT19606-r2 LB-WT19606-r3 LB-WT19606-r4 LB-Y1-r2 LB-Y1-r3 LB-Y1-r4 Mac-AB-r1 Mac-AB-r2 Mac-AB-r3 Mac-IJ-r1 Mac-IJ-r2 Mac-IJ-r4 Mac-W1-r1 Mac-W1-r2 Mac-W1-r3 Mac-WT19606-r2 Mac-WT19606-r3 Mac-WT19606-r4 Mac-Y1-r2 Mac-Y1-r3 Mac-Y1-r4; do java -jar /home/jhuang/Tools/Trimmomatic-0.36/trimmomatic-0.36.jar PE -threads 100 raw_data/${sample_id}_R1.fq.gz raw_data/${sample_id}_R2.fq.gz trimmed/${sample_id}_R1.fq.gz trimmed_unpaired/${sample_id}_R1.fq.gz trimmed/${sample_id}_R2.fq.gz trimmed_unpaired/${sample_id}_R2.fq.gz ILLUMINACLIP:/home/jhuang/Tools/Trimmomatic-0.36/adapters/TruSeq3-PE-2.fa:2:30:10:8:TRUE LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36 AVGQUAL:20; done 2> trimmomatic_pe.log; done -
Preparing samplesheet.csv
sample,fastq_1,fastq_2,strandedness LB-AB-r1,LB-AB-r1_R1.fq.gz,LB-AB-r1_R2.fq.gz,auto LB-AB-r2,LB-AB-r2_R1.fq.gz,LB-AB-r2_R2.fq.gz,auto LB-AB-r3,LB-AB-r3_R1.fq.gz,LB-AB-r3_R2.fq.gz,auto LB-IJ-r1,LB-IJ-r1_R1.fq.gz,LB-IJ-r1_R2.fq.gz,auto LB-IJ-r2,LB-IJ-r2_R1.fq.gz,LB-IJ-r2_R2.fq.gz,auto LB-IJ-r4,LB-IJ-r4_R1.fq.gz,LB-IJ-r4_R2.fq.gz,auto LB-W1-r1,LB-W1-r1_R1.fq.gz,LB-W1-r1_R2.fq.gz,auto LB-W1-r2,LB-W1-r2_R1.fq.gz,LB-W1-r2_R2.fq.gz,auto LB-W1-r3,LB-W1-r3_R1.fq.gz,LB-W1-r3_R2.fq.gz,auto LB-WT19606-r2,LB-WT19606-r2_R1.fq.gz,LB-WT19606-r2_R2.fq.gz,auto LB-WT19606-r3,LB-WT19606-r3_R1.fq.gz,LB-WT19606-r3_R2.fq.gz,auto LB-WT19606-r4,LB-WT19606-r4_R1.fq.gz,LB-WT19606-r4_R2.fq.gz,auto LB-Y1-r2,LB-Y1-r2_R1.fq.gz,LB-Y1-r2_R2.fq.gz,auto LB-Y1-r3,LB-Y1-r3_R1.fq.gz,LB-Y1-r3_R2.fq.gz,auto LB-Y1-r4,LB-Y1-r4_R1.fq.gz,LB-Y1-r4_R2.fq.gz,auto Mac-AB-r1,Mac-AB-r1_R1.fq.gz,Mac-AB-r1_R2.fq.gz,auto Mac-AB-r2,Mac-AB-r2_R1.fq.gz,Mac-AB-r2_R2.fq.gz,auto Mac-AB-r3,Mac-AB-r3_R1.fq.gz,Mac-AB-r3_R2.fq.gz,auto Mac-IJ-r1,Mac-IJ-r1_R1.fq.gz,Mac-IJ-r1_R2.fq.gz,auto Mac-IJ-r2,Mac-IJ-r2_R1.fq.gz,Mac-IJ-r2_R2.fq.gz,auto Mac-IJ-r4,Mac-IJ-r4_R1.fq.gz,Mac-IJ-r4_R2.fq.gz,auto Mac-W1-r1,Mac-W1-r1_R1.fq.gz,Mac-W1-r1_R2.fq.gz,auto Mac-W1-r2,Mac-W1-r2_R1.fq.gz,Mac-W1-r2_R2.fq.gz,auto Mac-W1-r3,Mac-W1-r3_R1.fq.gz,Mac-W1-r3_R2.fq.gz,auto Mac-WT19606-r2,Mac-WT19606-r2_R1.fq.gz,Mac-WT19606-r2_R2.fq.gz,auto Mac-WT19606-r3,Mac-WT19606-r3_R1.fq.gz,Mac-WT19606-r3_R2.fq.gz,auto Mac-WT19606-r4,Mac-WT19606-r4_R1.fq.gz,Mac-WT19606-r4_R2.fq.gz,auto Mac-Y1-r2,Mac-Y1-r2_R1.fq.gz,Mac-Y1-r2_R2.fq.gz,auto Mac-Y1-r3,Mac-Y1-r3_R1.fq.gz,Mac-Y1-r3_R2.fq.gz,auto Mac-Y1-r4,Mac-Y1-r4_R1.fq.gz,Mac-Y1-r4_R2.fq.gz,auto #mv trimmed/* . -
nextflow run
#Example1: http://xgenes.com/article/article-content/157/prepare-virus-gtf-for-nextflow-run/ #docker pull nfcore/rnaseq ln -s /home/jhuang/Tools/nf-core-rnaseq-3.12.0/ rnaseq # ---- SUCCESSFUL with directly downloaded gff3 and fasta from NCBI using docker after replacing 'CDS' with 'exon' ---- (host_env) /usr/local/bin/nextflow run rnaseq/main.nf --input samplesheet.csv --outdir results --fasta "/home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040.fasta" --gff "/home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_m.gff" -profile docker -resume --max_cpus 55 --max_memory 512.GB --max_time 2400.h --save_align_intermeds --save_unaligned --save_reference --aligner 'star_salmon' --gtf_group_features 'gene_id' --gtf_extra_attributes 'gene_name' --featurecounts_group_type 'gene_biotype' --featurecounts_feature_type 'transcript' -
Import data and pca-plot
#mamba activate r_env #install.packages("ggfun") # Import the required libraries library("AnnotationDbi") library("clusterProfiler") library("ReactomePA") library(gplots) library(tximport) library(DESeq2) #library("org.Hs.eg.db") library(dplyr) library(tidyverse) #install.packages("devtools") #devtools::install_version("gtable", version = "0.3.0") library(gplots) library("RColorBrewer") #install.packages("ggrepel") library("ggrepel") # install.packages("openxlsx") library(openxlsx) library(EnhancedVolcano) library(DESeq2) library(edgeR) setwd("~/DATA/Data_Tam_RNAseq_2025_LB_vs_Mac_ATCC19606/results/star_salmon") # Define paths to your Salmon output quantification files files <- c("LB-AB_r1" = "./LB-AB-r1/quant.sf", "LB-AB_r2" = "./LB-AB-r2/quant.sf", "LB-AB_r3" = "./LB-AB-r3/quant.sf", "LB-IJ_r1" = "./LB-IJ-r1/quant.sf", "LB-IJ_r2" = "./LB-IJ-r2/quant.sf", "LB-IJ_r4" = "./LB-IJ-r4/quant.sf", "LB-W1_r1" = "./LB-W1-r1/quant.sf", "LB-W1_r2" = "./LB-W1-r2/quant.sf", "LB-W1_r3" = "./LB-W1-r3/quant.sf", "LB-WT19606_r2" = "./LB-WT19606-r2/quant.sf", "LB-WT19606_r3" = "./LB-WT19606-r3/quant.sf", "LB-WT19606_r4" = "./LB-WT19606-r4/quant.sf", "LB-Y1_r2" = "./LB-Y1-r2/quant.sf", "LB-Y1_r3" = "./LB-Y1-r3/quant.sf", "LB-Y1_r4" = "./LB-Y1-r4/quant.sf", "Mac-AB_r1" = "./Mac-AB-r1/quant.sf", "Mac-AB_r2" = "./Mac-AB-r2/quant.sf", "Mac-AB_r3" = "./Mac-AB-r3/quant.sf", "Mac-IJ_r1" = "./Mac-IJ-r1/quant.sf", "Mac-IJ_r2" = "./Mac-IJ-r2/quant.sf", "Mac-IJ_r4" = "./Mac-IJ-r4/quant.sf", "Mac-W1_r1" = "./Mac-W1-r1/quant.sf", "Mac-W1_r2" = "./Mac-W1-r2/quant.sf", "Mac-W1_r3" = "./Mac-W1-r3/quant.sf", "Mac-WT19606_r2" = "./Mac-WT19606-r2/quant.sf", "Mac-WT19606_r3" = "./Mac-WT19606-r3/quant.sf", "Mac-WT19606_r4" = "./Mac-WT19606-r4/quant.sf", "Mac-Y1_r2" = "./Mac-Y1-r2/quant.sf", "Mac-Y1_r3" = "./Mac-Y1-r3/quant.sf", "Mac-Y1_r4" = "./Mac-Y1-r4/quant.sf") # Import the transcript abundance data with tximport txi <- tximport(files, type = "salmon", txIn = TRUE, txOut = TRUE) # Define the replicates and condition of the samples #replicate <- factor(c("r1", "r2", "r3", "r1", "r2", "r3", "r1", "r2", "r3")) #adeA and adeB encode a membrane fusion protein that is part of the AdeABC efflux pump, which contributes to multidrug resistance. #System: Part of the AdeIJK efflux pump, which includes: adeI — membrane fusion protein, adeJ — RND transporter, adeK — outer membrane factor condition <- factor(c("LB-AB","LB-AB","LB-AB", "LB-IJ","LB-IJ","LB-IJ", "LB-W1","LB-W1","LB-W1","LB-WT19606","LB-WT19606","LB-WT19606","LB-Y1","LB-Y1","LB-Y1","Mac-AB","Mac-AB","Mac-AB","Mac-IJ","Mac-IJ","Mac-IJ","Mac-W1","Mac-W1","Mac-W1","Mac-WT19606","Mac-WT19606","Mac-WT19606","Mac-Y1","Mac-Y1","Mac-Y1")) # Define the colData for DESeq2 colData <- data.frame(condition=condition, row.names=names(files)) # ------------------------ # 1️⃣ Setup and input files # ------------------------ # Read in transcript-to-gene mapping tx2gene <- read.table("salmon_tx2gene.tsv", header=FALSE, stringsAsFactors=FALSE) colnames(tx2gene) <- c("transcript_id", "gene_id", "gene_name") # Prepare tx2gene for gene-level summarization (remove gene_name if needed) tx2gene_geneonly <- tx2gene[, c("transcript_id", "gene_id")] # ------------------------------- # 2️⃣ Transcript-level counts # ------------------------------- # Create DESeqDataSet directly from tximport (transcript-level) dds_tx <- DESeqDataSetFromTximport(txi, colData=colData, design=~condition) write.csv(counts(dds_tx), file="transcript_counts.csv") # -------------------------------- # 3️⃣ Gene-level summarization # -------------------------------- # Re-import Salmon data summarized at gene level txi_gene <- tximport(files, type="salmon", tx2gene=tx2gene_geneonly, txOut=FALSE) # Create DESeqDataSet for gene-level counts #dds <- DESeqDataSetFromTximport(txi_gene, colData=colData, design=~condition+replicate) dds <- DESeqDataSetFromTximport(txi_gene, colData=colData, design=~condition) # -------------------------------- # 4️⃣ Raw counts table (with gene names) # -------------------------------- # Extract raw gene-level counts counts_data <- as.data.frame(counts(dds, normalized=FALSE)) counts_data$gene_id <- rownames(counts_data) # Add gene names tx2gene_unique <- unique(tx2gene[, c("gene_id", "gene_name")]) counts_data <- merge(counts_data, tx2gene_unique, by="gene_id", all.x=TRUE) # Reorder columns: gene_id, gene_name, then counts count_cols <- setdiff(colnames(counts_data), c("gene_id", "gene_name")) counts_data <- counts_data[, c("gene_id", "gene_name", count_cols)] # -------------------------------- # 5️⃣ Calculate CPM # -------------------------------- library(edgeR) library(openxlsx) # Prepare count matrix for CPM calculation count_matrix <- as.matrix(counts_data[, !(colnames(counts_data) %in% c("gene_id", "gene_name"))]) # Calculate CPM #cpm_matrix <- cpm(count_matrix, normalized.lib.sizes=FALSE) total_counts <- colSums(count_matrix) cpm_matrix <- t(t(count_matrix) / total_counts) * 1e6 cpm_matrix <- as.data.frame(cpm_matrix) # Add gene_id and gene_name back to CPM table cpm_counts <- cbind(counts_data[, c("gene_id", "gene_name")], cpm_matrix) # -------------------------------- # 6️⃣ Save outputs # -------------------------------- write.csv(counts_data, "gene_raw_counts.csv", row.names=FALSE) write.xlsx(counts_data, "gene_raw_counts.xlsx", row.names=FALSE) write.xlsx(cpm_counts, "gene_cpm_counts.xlsx", row.names=FALSE) # -- (Optional) Save the rlog-transformed counts -- dim(counts(dds)) head(counts(dds), 10) rld <- rlogTransformation(dds) rlog_counts <- assay(rld) write.xlsx(as.data.frame(rlog_counts), "gene_rlog_transformed_counts.xlsx") # ---- (Optional for NACHREIHEN) split the factos media and strain from condition (for comparison Mac vs LB) ---- # AdeIJK vs. AdeABC Efflux Pumps # * AdeIJK is the "housekeeping" pump — always active, broadly expressed, contributing to background resistance. # * AdeABC is the "emergency" pump — induced under stress or mutations, more potent in contributing to clinical multidrug resistance. #LB = Luria-Bertani broth (a standard rich growth medium) #Mac = MacConkey agar or broth (selective for Gram-negative bacteria) # - Growth medium Media or Condition, GrowthMedium # - Bacterial strain/genotype Strain or Isolate, Genotype, SampleType media <- factor(c("LB","LB","LB", "LB","LB","LB", "LB","LB","LB","LB","LB","LB","LB","LB","LB","Mac","Mac","Mac","Mac","Mac","Mac","Mac","Mac","Mac","Mac","Mac","Mac","Mac","Mac","Mac")) strain <- factor(c("AB","AB","AB", "IJ","IJ","IJ", "W1","W1","W1","WT19606","WT19606","WT19606","Y1","Y1","Y1","AB","AB","AB","IJ","IJ","IJ","W1","W1","W1","WT19606","WT19606","WT19606","Y1","Y1","Y1")) # Define the colData for DESeq2 colData <- data.frame(media=media, strain=strain, row.names=names(files)) # -- transcript-level count data (x2) -- # Create DESeqDataSet object dds <- DESeqDataSetFromTximport(txi, colData=colData, design=~media+strain) #write.csv(counts(dds), file="transcript_counts_media_strain.csv") #check correctness, it should be identical to transcript_counts.csv # -- gene-level count data (x2) -- # Read in the tx2gene map from salmon_tx2gene.tsv tx2gene <- read.table("salmon_tx2gene.tsv", header=FALSE, stringsAsFactors=FALSE) # Set the column names colnames(tx2gene) <- c("transcript_id", "gene_id", "gene_name") # Remove the gene_name column if not needed tx2gene <- tx2gene[,1:2] # Import and summarize the Salmon data with tximport txi <- tximport(files, type = "salmon", tx2gene = tx2gene, txOut = FALSE) # Continue with the DESeq2 workflow as before... colData <- data.frame(media=media, strain=strain, row.names=names(files)) dds <- DESeqDataSetFromTximport(txi, colData=colData, design=~media+strain) #dds <- dds[rowSums(counts(dds) > 3) > 2, ] #3796->???? #write.csv(counts(dds, normalized=FALSE), file="gene_counts_media_strain.csv") #check correctness, it should be identical to gene_counts.csv # ---- (Optional for NACHREIHEN) END ---- # -- pca -- png("pca2.png", 1200, 800) plotPCA(rld, intgroup=c("condition")) dev.off() # -- heatmap -- png("heatmap2.png", 1200, 800) distsRL <- dist(t(assay(rld))) mat <- as.matrix(distsRL) hc <- hclust(distsRL) hmcol <- colorRampPalette(brewer.pal(9,"GnBu"))(100) heatmap.2(mat, Rowv=as.dendrogram(hc),symm=TRUE, trace="none",col = rev(hmcol), margin=c(13, 13)) dev.off() # -- pca_media_strain -- png("pca_media.png", 1200, 800) plotPCA(rld, intgroup=c("media")) dev.off() png("pca_strain.png", 1200, 800) plotPCA(rld, intgroup=c("strain")) dev.off() -
(Optional; ERROR-->need to be debugged!) ) estimate size factors and dispersion values.
#Size Factors: These are used to normalize the read counts across different samples. The size factor for a sample accounts for differences in sequencing depth (i.e., the total number of reads) and other technical biases between samples. After normalization with size factors, the counts should be comparable across samples. Size factors are usually calculated in a way that they reflect the median or mean ratio of gene expression levels between samples, assuming that most genes are not differentially expressed. #Dispersion: This refers to the variability or spread of gene expression measurements. In RNA-seq data analysis, each gene has its own dispersion value, which reflects how much the counts for that gene vary between different samples, more than what would be expected just due to the Poisson variation inherent in counting. Dispersion is important for accurately modeling the data and for detecting differentially expressed genes. #So in summary, size factors are specific to samples (used to make counts comparable across samples), and dispersion values are specific to genes (reflecting variability in gene expression). sizeFactors(dds) #NULL # Estimate size factors dds <- estimateSizeFactors(dds) # Estimate dispersions dds <- estimateDispersions(dds) #> sizeFactors(dds) #control_r1 control_r2 HSV.d2_r1 HSV.d2_r2 HSV.d4_r1 HSV.d4_r2 HSV.d6_r1 #2.3282468 2.0251928 1.8036883 1.3767551 0.9341929 1.0911693 0.5454526 #HSV.d6_r2 HSV.d8_r1 HSV.d8_r2 #0.4604461 0.5799834 0.6803681 # (DEBUG) If avgTxLength is Necessary #To simplify the computation and ensure sizeFactors are calculated: assays(dds)$avgTxLength <- NULL dds <- estimateSizeFactors(dds) sizeFactors(dds) #If you want to retain avgTxLength but suspect it is causing issues, you can explicitly instruct DESeq2 to compute size factors without correcting for library size with average transcript lengths: dds <- estimateSizeFactors(dds, controlGenes = NULL, use = FALSE) sizeFactors(dds) # If alone with virus data, the following BUG occured: #Still NULL --> BUG --> using manual calculation method for sizeFactor calculation! HeLa_TO_r1 HeLa_TO_r2 0.9978755 1.1092227 data.frame(genes = rownames(dds), dispersions = dispersions(dds)) #Given the raw counts, the control_r1 and control_r2 samples seem to have a much lower sequencing depth (total read count) than the other samples. Therefore, when normalization methods are applied, the normalization factors for these control samples will be relatively high, boosting the normalized counts. 1/0.9978755=1.002129023 1/1.1092227= #bamCoverage --bam ../markDuplicates/${sample}Aligned.sortedByCoord.out.bam -o ${sample}_norm.bw --binSize 10 --scaleFactor --effectiveGenomeSize 2864785220 bamCoverage --bam ../markDuplicates/HeLa_TO_r1Aligned.sortedByCoord.out.markDups.bam -o HeLa_TO_r1.bw --binSize 10 --scaleFactor 1.002129023 --effectiveGenomeSize 2864785220 bamCoverage --bam ../markDuplicates/HeLa_TO_r2Aligned.sortedByCoord.out.markDups.bam -o HeLa_TO_r2.bw --binSize 10 --scaleFactor 0.901532217 --effectiveGenomeSize 2864785220 raw_counts <- counts(dds) normalized_counts <- counts(dds, normalized=TRUE) #write.table(raw_counts, file="raw_counts.txt", sep="\t", quote=F, col.names=NA) #write.table(normalized_counts, file="normalized_counts.txt", sep="\t", quote=F, col.names=NA) #convert bam to bigwig using deepTools by feeding inverse of DESeq’s size Factor estimSf <- function (cds){ # Get the count matrix cts <- counts(cds) # Compute the geometric mean geomMean <- function(x) prod(x)^(1/length(x)) # Compute the geometric mean over the line gm.mean <- apply(cts, 1, geomMean) # Zero values are set to NA (avoid subsequentcdsdivision by 0) gm.mean[gm.mean == 0] <- NA # Divide each line by its corresponding geometric mean # sweep(x, MARGIN, STATS, FUN = "-", check.margin = TRUE, ...) # MARGIN: 1 or 2 (line or columns) # STATS: a vector of length nrow(x) or ncol(x), depending on MARGIN # FUN: the function to be applied cts <- sweep(cts, 1, gm.mean, FUN="/") # Compute the median over the columns med <- apply(cts, 2, median, na.rm=TRUE) # Return the scaling factor return(med) } #https://dputhier.github.io/ASG/practicals/rnaseq_diff_Snf2/rnaseq_diff_Snf2.html #http://bioconductor.org/packages/devel/bioc/vignettes/DESeq2/inst/doc/DESeq2.html#data-transformations-and-visualization #https://hbctraining.github.io/DGE_workshop/lessons/02_DGE_count_normalization.html #https://hbctraining.github.io/DGE_workshop/lessons/04_DGE_DESeq2_analysis.html #https://genviz.org/module-04-expression/0004/02/01/DifferentialExpression/ #DESeq2’s median of ratios [1] #EdgeR’s trimmed mean of M values (TMM) [2] #http://www.nathalievialaneix.eu/doc/html/TP1_normalization.html #very good website! test_normcount <- sweep(raw_counts, 2, sizeFactors(dds), "/") sum(test_normcount != normalized_counts) -
Select the differentially expressed genes
#https://galaxyproject.eu/posts/2020/08/22/three-steps-to-galaxify-your-tool/ #https://www.biostars.org/p/282295/ #https://www.biostars.org/p/335751/ #> dds$condition #LB-AB LB-IJ LB-W1 LB-WT19606 LB-Y1 Mac-AB Mac-IJ Mac-W1 Mac-WT19606 Mac-Y1 #CONSOLE: mkdir star_salmon/degenes setwd("degenes") #---- relevel to control ---- dds$condition <- relevel(dds$condition, "LB-WT19606") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("LB.AB_vs_LB.WT19606","LB.IJ_vs_LB.WT19606","LB.W1_vs_LB.WT19606","LB.Y1_vs_LB.WT19606") dds$condition <- relevel(dds$condition, "Mac-WT19606") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("Mac.AB_vs_Mac.WT19606","Mac.IJ_vs_Mac.WT19606","Mac.W1_vs_Mac.WT19606","Mac.Y1_vs_Mac.WT19606") # - 如果你的实验是关注细菌在没有选择性压力下的生长、基因表达或一般行为,LB 是更好的对照。 # - 如果你希望研究细菌在选择性压力下的行为(例如,针对革兰氏阴性细菌、测试抗生素耐药性或区分乳糖发酵菌),那么 MacConkey 更适合作为对照。 dds$media <- relevel(dds$media, "LB") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("Mac_vs_LB") dds$media <- relevel(dds$media, "Mac") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("LB_vs_Mac") for (i in clist) { #contrast = paste("condition", i, sep="_") contrast = paste("media", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) } # -- Under host-env -- grep -P "\tgene\t" CP059040.gff > CP059040_gene.gff python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff LB.AB_vs_LB.WT19606-all.txt LB.AB_vs_LB.WT19606-all.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff LB.AB_vs_LB.WT19606-up.txt LB.AB_vs_LB.WT19606-up.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff LB.AB_vs_LB.WT19606-down.txt LB.AB_vs_LB.WT19606-down.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff LB.IJ_vs_LB.WT19606-all.txt LB.IJ_vs_LB.WT19606-all.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff LB.IJ_vs_LB.WT19606-up.txt LB.IJ_vs_LB.WT19606-up.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff LB.IJ_vs_LB.WT19606-down.txt LB.IJ_vs_LB.WT19606-down.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff LB.W1_vs_LB.WT19606-all.txt LB.W1_vs_LB.WT19606-all.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff LB.W1_vs_LB.WT19606-up.txt LB.W1_vs_LB.WT19606-up.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff LB.W1_vs_LB.WT19606-down.txt LB.W1_vs_LB.WT19606-down.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff LB.Y1_vs_LB.WT19606-all.txt LB.Y1_vs_LB.WT19606-all.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff LB.Y1_vs_LB.WT19606-up.txt LB.Y1_vs_LB.WT19606-up.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff LB.Y1_vs_LB.WT19606-down.txt LB.Y1_vs_LB.WT19606-down.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff Mac.AB_vs_Mac.WT19606-all.txt Mac.AB_vs_Mac.WT19606-all.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff Mac.AB_vs_Mac.WT19606-up.txt Mac.AB_vs_Mac.WT19606-up.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff Mac.AB_vs_Mac.WT19606-down.txt Mac.AB_vs_Mac.WT19606-down.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff Mac.IJ_vs_Mac.WT19606-all.txt Mac.IJ_vs_Mac.WT19606-all.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff Mac.IJ_vs_Mac.WT19606-up.txt Mac.IJ_vs_Mac.WT19606-up.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff Mac.IJ_vs_Mac.WT19606-down.txt Mac.IJ_vs_Mac.WT19606-down.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff Mac.W1_vs_Mac.WT19606-all.txt Mac.W1_vs_Mac.WT19606-all.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff Mac.W1_vs_Mac.WT19606-up.txt Mac.W1_vs_Mac.WT19606-up.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff Mac.W1_vs_Mac.WT19606-down.txt Mac.W1_vs_Mac.WT19606-down.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff Mac.Y1_vs_Mac.WT19606-all.txt Mac.Y1_vs_Mac.WT19606-all.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff Mac.Y1_vs_Mac.WT19606-up.txt Mac.Y1_vs_Mac.WT19606-up.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff Mac.Y1_vs_Mac.WT19606-down.txt Mac.Y1_vs_Mac.WT19606-down.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff Mac_vs_LB-all.txt Mac_vs_LB-all.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff Mac_vs_LB-up.txt Mac_vs_LB-up.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff Mac_vs_LB-down.txt Mac_vs_LB-down.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff LB_vs_Mac-all.txt LB_vs_Mac-all.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff LB_vs_Mac-up.txt LB_vs_Mac-up.csv python3 ~/Scripts/replace_gene_names.py /home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_gene.gff LB_vs_Mac-down.txt LB_vs_Mac-down.csv # ---- Mac_vs_LB ---- res <- read.csv("Mac_vs_LB-all.csv") # Replace empty GeneName with modified GeneID res$GeneName <- ifelse( res$GeneName == "" | is.na(res$GeneName), gsub("gene-", "", res$GeneID), res$GeneName ) duplicated_genes <- res[duplicated(res$GeneName), "GeneName"] #print(duplicated_genes) # [1] "bfr" "lipA" "ahpF" "pcaF" "alr" "pcaD" "cydB" "lpdA" "pgaC" "ppk1" #[11] "pcaF" "tuf" "galE" "murI" "yccS" "rrf" "rrf" "arsB" "ptsP" "umuD" #[21] "map" "pgaB" "rrf" "rrf" "rrf" "pgaD" "uraH" "benE" #res[res$GeneName == "bfr", ] #1st_strategy First occurrence is kept and Subsequent duplicates are removed #res <- res[!duplicated(res$GeneName), ] #2nd_strategy keep the row with the smallest padj value for each GeneName res <- res %>% group_by(GeneName) %>% slice_min(padj, with_ties = FALSE) %>% ungroup() res <- as.data.frame(res) # Sort res first by padj (ascending) and then by log2FoldChange (descending) res <- res[order(res$padj, -res$log2FoldChange), ] # Assuming res is your dataframe and already processed # Filter up-regulated genes: log2FoldChange > 2 and padj < 1e-2 up_regulated <- res[res$log2FoldChange > 2 & res$padj < 1e-2, ] # Filter down-regulated genes: log2FoldChange < -2 and padj < 1e-2 down_regulated <- res[res$log2FoldChange < -2 & res$padj < 1e-2, ] # Create a new workbook wb <- createWorkbook() # Add the complete dataset as the first sheet addWorksheet(wb, "Complete_Data") writeData(wb, "Complete_Data", res) # Add the up-regulated genes as the second sheet addWorksheet(wb, "Up_Regulated") writeData(wb, "Up_Regulated", up_regulated) # Add the down-regulated genes as the third sheet addWorksheet(wb, "Down_Regulated") writeData(wb, "Down_Regulated", down_regulated) # Save the workbook to a file saveWorkbook(wb, "Gene_Expression_Mac_vs_LB.xlsx", overwrite = TRUE) # Set the 'GeneName' column as row.names rownames(res) <- res$GeneName # Drop the 'GeneName' column since it's now the row names res$GeneName <- NULL head(res) ## Ensure the data frame matches the expected format ## For example, it should have columns: log2FoldChange, padj, etc. #res <- as.data.frame(res) ## Remove rows with NA in log2FoldChange (if needed) #res <- res[!is.na(res$log2FoldChange),] # Replace padj = 0 with a small value res$padj[res$padj == 0] <- 1e-150 #library(EnhancedVolcano) # Assuming res is already sorted and processed png("Mac_vs_LB.png", width=1200, height=2000) #max.overlaps = 10 EnhancedVolcano(res, lab = rownames(res), x = 'log2FoldChange', y = 'padj', pCutoff = 1e-2, FCcutoff = 2, title = '', subtitleLabSize = 18, pointSize = 3.0, labSize = 5.0, colAlpha = 1, legendIconSize = 4.0, drawConnectors = TRUE, widthConnectors = 0.5, colConnectors = 'black', subtitle = expression("Mac versus LB")) dev.off() # ---- LB.AB_vs_LB.WT19606 ---- res <- read.csv("LB.AB_vs_LB.WT19606-all.csv") # Replace empty GeneName with modified GeneID res$GeneName <- ifelse( res$GeneName == "" | is.na(res$GeneName), gsub("gene-", "", res$GeneID), res$GeneName ) duplicated_genes <- res[duplicated(res$GeneName), "GeneName"] res <- res %>% group_by(GeneName) %>% slice_min(padj, with_ties = FALSE) %>% ungroup() res <- as.data.frame(res) # Sort res first by padj (ascending) and then by log2FoldChange (descending) res <- res[order(res$padj, -res$log2FoldChange), ] # Assuming res is your dataframe and already processed # Filter up-regulated genes: log2FoldChange > 2 and padj < 1e-2 up_regulated <- res[res$log2FoldChange > 2 & res$padj < 1e-2, ] # Filter down-regulated genes: log2FoldChange < -2 and padj < 1e-2 down_regulated <- res[res$log2FoldChange < -2 & res$padj < 1e-2, ] # Create a new workbook wb <- createWorkbook() # Add the complete dataset as the first sheet addWorksheet(wb, "Complete_Data") writeData(wb, "Complete_Data", res) # Add the up-regulated genes as the second sheet addWorksheet(wb, "Up_Regulated") writeData(wb, "Up_Regulated", up_regulated) # Add the down-regulated genes as the third sheet addWorksheet(wb, "Down_Regulated") writeData(wb, "Down_Regulated", down_regulated) # Save the workbook to a file saveWorkbook(wb, "Gene_Expression_LB.AB_vs_LB.WT19606.xlsx", overwrite = TRUE) # Set the 'GeneName' column as row.names rownames(res) <- res$GeneName # Drop the 'GeneName' column since it's now the row names res$GeneName <- NULL head(res) ## Ensure the data frame matches the expected format ## For example, it should have columns: log2FoldChange, padj, etc. #res <- as.data.frame(res) ## Remove rows with NA in log2FoldChange (if needed) #res <- res[!is.na(res$log2FoldChange),] # Replace padj = 0 with a small value res$padj[res$padj == 0] <- 1e-12 #library(EnhancedVolcano) # Assuming res is already sorted and processed png("LB.AB_vs_LB.WT19606.png", width=1200, height=1200) #max.overlaps = 10 EnhancedVolcano(res, lab = rownames(res), x = 'log2FoldChange', y = 'padj', pCutoff = 1e-2, FCcutoff = 2, title = '', subtitleLabSize = 18, pointSize = 3.0, labSize = 5.0, colAlpha = 1, legendIconSize = 4.0, drawConnectors = TRUE, widthConnectors = 0.5, colConnectors = 'black', subtitle = expression("LB.AB versus LB.WT19606")) dev.off() # ---- LB.IJ_vs_LB.WT19606 ---- res <- read.csv("LB.IJ_vs_LB.WT19606-all.csv") # Replace empty GeneName with modified GeneID res$GeneName <- ifelse( res$GeneName == "" | is.na(res$GeneName), gsub("gene-", "", res$GeneID), res$GeneName ) duplicated_genes <- res[duplicated(res$GeneName), "GeneName"] res <- res %>% group_by(GeneName) %>% slice_min(padj, with_ties = FALSE) %>% ungroup() res <- as.data.frame(res) # Sort res first by padj (ascending) and then by log2FoldChange (descending) res <- res[order(res$padj, -res$log2FoldChange), ] # Assuming res is your dataframe and already processed # Filter up-regulated genes: log2FoldChange > 2 and padj < 1e-2 up_regulated <- res[res$log2FoldChange > 2 & res$padj < 1e-2, ] # Filter down-regulated genes: log2FoldChange < -2 and padj < 1e-2 down_regulated <- res[res$log2FoldChange < -2 & res$padj < 1e-2, ] # Create a new workbook wb <- createWorkbook() # Add the complete dataset as the first sheet addWorksheet(wb, "Complete_Data") writeData(wb, "Complete_Data", res) # Add the up-regulated genes as the second sheet addWorksheet(wb, "Up_Regulated") writeData(wb, "Up_Regulated", up_regulated) # Add the down-regulated genes as the third sheet addWorksheet(wb, "Down_Regulated") writeData(wb, "Down_Regulated", down_regulated) # Save the workbook to a file saveWorkbook(wb, "Gene_Expression_LB.IJ_vs_LB.WT19606.xlsx", overwrite = TRUE) # Set the 'GeneName' column as row.names rownames(res) <- res$GeneName # Drop the 'GeneName' column since it's now the row names res$GeneName <- NULL head(res) ## Ensure the data frame matches the expected format ## For example, it should have columns: log2FoldChange, padj, etc. #res <- as.data.frame(res) ## Remove rows with NA in log2FoldChange (if needed) #res <- res[!is.na(res$log2FoldChange),] # Replace padj = 0 with a small value res$padj[res$padj == 0] <- 1e-12 #library(EnhancedVolcano) # Assuming res is already sorted and processed png("LB.IJ_vs_LB.WT19606.png", width=1200, height=1200) #max.overlaps = 10 EnhancedVolcano(res, lab = rownames(res), x = 'log2FoldChange', y = 'padj', pCutoff = 1e-2, FCcutoff = 2, title = '', subtitleLabSize = 18, pointSize = 3.0, labSize = 5.0, colAlpha = 1, legendIconSize = 4.0, drawConnectors = TRUE, widthConnectors = 0.5, colConnectors = 'black', subtitle = expression("LB.IJ versus LB.WT19606")) dev.off() # ---- LB.W1_vs_LB.WT19606 ---- res <- read.csv("LB.W1_vs_LB.WT19606-all.csv") # Replace empty GeneName with modified GeneID res$GeneName <- ifelse( res$GeneName == "" | is.na(res$GeneName), gsub("gene-", "", res$GeneID), res$GeneName ) duplicated_genes <- res[duplicated(res$GeneName), "GeneName"] res <- res %>% group_by(GeneName) %>% slice_min(padj, with_ties = FALSE) %>% ungroup() res <- as.data.frame(res) # Sort res first by padj (ascending) and then by log2FoldChange (descending) res <- res[order(res$padj, -res$log2FoldChange), ] # Assuming res is your dataframe and already processed # Filter up-regulated genes: log2FoldChange > 2 and padj < 1e-2 up_regulated <- res[res$log2FoldChange > 2 & res$padj < 1e-2, ] # Filter down-regulated genes: log2FoldChange < -2 and padj < 1e-2 down_regulated <- res[res$log2FoldChange < -2 & res$padj < 1e-2, ] # Create a new workbook wb <- createWorkbook() # Add the complete dataset as the first sheet addWorksheet(wb, "Complete_Data") writeData(wb, "Complete_Data", res) # Add the up-regulated genes as the second sheet addWorksheet(wb, "Up_Regulated") writeData(wb, "Up_Regulated", up_regulated) # Add the down-regulated genes as the third sheet addWorksheet(wb, "Down_Regulated") writeData(wb, "Down_Regulated", down_regulated) # Save the workbook to a file saveWorkbook(wb, "Gene_Expression_LB.W1_vs_LB.WT19606.xlsx", overwrite = TRUE) # Set the 'GeneName' column as row.names rownames(res) <- res$GeneName # Drop the 'GeneName' column since it's now the row names res$GeneName <- NULL head(res) ## Ensure the data frame matches the expected format ## For example, it should have columns: log2FoldChange, padj, etc. #res <- as.data.frame(res) ## Remove rows with NA in log2FoldChange (if needed) #res <- res[!is.na(res$log2FoldChange),] # Replace padj = 0 with a small value res$padj[res$padj == 0] <- 1e-12 #library(EnhancedVolcano) # Assuming res is already sorted and processed png("LB.W1_vs_LB.WT19606.png", width=1200, height=1200) #max.overlaps = 10 EnhancedVolcano(res, lab = rownames(res), x = 'log2FoldChange', y = 'padj', pCutoff = 1e-2, FCcutoff = 2, title = '', subtitleLabSize = 18, pointSize = 3.0, labSize = 5.0, colAlpha = 1, legendIconSize = 4.0, drawConnectors = TRUE, widthConnectors = 0.5, colConnectors = 'black', subtitle = expression("LB.W1 versus LB.WT19606")) dev.off() # ---- LB.Y1_vs_LB.WT19606 ---- res <- read.csv("LB.Y1_vs_LB.WT19606-all.csv") # Replace empty GeneName with modified GeneID res$GeneName <- ifelse( res$GeneName == "" | is.na(res$GeneName), gsub("gene-", "", res$GeneID), res$GeneName ) duplicated_genes <- res[duplicated(res$GeneName), "GeneName"] res <- res %>% group_by(GeneName) %>% slice_min(padj, with_ties = FALSE) %>% ungroup() res <- as.data.frame(res) # Sort res first by padj (ascending) and then by log2FoldChange (descending) res <- res[order(res$padj, -res$log2FoldChange), ] # Assuming res is your dataframe and already processed # Filter up-regulated genes: log2FoldChange > 2 and padj < 1e-2 up_regulated <- res[res$log2FoldChange > 2 & res$padj < 1e-2, ] # Filter down-regulated genes: log2FoldChange < -2 and padj < 1e-2 down_regulated <- res[res$log2FoldChange < -2 & res$padj < 1e-2, ] # Create a new workbook wb <- createWorkbook() # Add the complete dataset as the first sheet addWorksheet(wb, "Complete_Data") writeData(wb, "Complete_Data", res) # Add the up-regulated genes as the second sheet addWorksheet(wb, "Up_Regulated") writeData(wb, "Up_Regulated", up_regulated) # Add the down-regulated genes as the third sheet addWorksheet(wb, "Down_Regulated") writeData(wb, "Down_Regulated", down_regulated) # Save the workbook to a file saveWorkbook(wb, "Gene_Expression_LB.Y1_vs_LB.WT19606.xlsx", overwrite = TRUE) # Set the 'GeneName' column as row.names rownames(res) <- res$GeneName # Drop the 'GeneName' column since it's now the row names res$GeneName <- NULL head(res) ## Ensure the data frame matches the expected format ## For example, it should have columns: log2FoldChange, padj, etc. #res <- as.data.frame(res) ## Remove rows with NA in log2FoldChange (if needed) #res <- res[!is.na(res$log2FoldChange),] # Replace padj = 0 with a small value res$padj[res$padj == 0] <- 1e-12 #library(EnhancedVolcano) # Assuming res is already sorted and processed png("LB.Y1_vs_LB.WT19606.png", width=1200, height=1200) #max.overlaps = 10 EnhancedVolcano(res, lab = rownames(res), x = 'log2FoldChange', y = 'padj', pCutoff = 1e-2, FCcutoff = 2, title = '', subtitleLabSize = 18, pointSize = 3.0, labSize = 5.0, colAlpha = 1, legendIconSize = 4.0, drawConnectors = TRUE, widthConnectors = 0.5, colConnectors = 'black', subtitle = expression("LB.Y1 versus LB.WT19606")) dev.off() # ---- Mac.AB_vs_Mac.WT19606 ---- res <- read.csv("Mac.AB_vs_Mac.WT19606-all.csv") # Replace empty GeneName with modified GeneID res$GeneName <- ifelse( res$GeneName == "" | is.na(res$GeneName), gsub("gene-", "", res$GeneID), res$GeneName ) duplicated_genes <- res[duplicated(res$GeneName), "GeneName"] res <- res %>% group_by(GeneName) %>% slice_min(padj, with_ties = FALSE) %>% ungroup() res <- as.data.frame(res) # Sort res first by padj (ascending) and then by log2FoldChange (descending) res <- res[order(res$padj, -res$log2FoldChange), ] # Assuming res is your dataframe and already processed # Filter up-regulated genes: log2FoldChange > 2 and padj < 1e-2 up_regulated <- res[res$log2FoldChange > 2 & res$padj < 1e-2, ] # Filter down-regulated genes: log2FoldChange < -2 and padj < 1e-2 down_regulated <- res[res$log2FoldChange < -2 & res$padj < 1e-2, ] # Create a new workbook wb <- createWorkbook() # Add the complete dataset as the first sheet addWorksheet(wb, "Complete_Data") writeData(wb, "Complete_Data", res) # Add the up-regulated genes as the second sheet addWorksheet(wb, "Up_Regulated") writeData(wb, "Up_Regulated", up_regulated) # Add the down-regulated genes as the third sheet addWorksheet(wb, "Down_Regulated") writeData(wb, "Down_Regulated", down_regulated) # Save the workbook to a file saveWorkbook(wb, "Gene_Expression_Mac.AB_vs_Mac.WT19606.xlsx", overwrite = TRUE) # Set the 'GeneName' column as row.names rownames(res) <- res$GeneName # Drop the 'GeneName' column since it's now the row names res$GeneName <- NULL head(res) ## Ensure the data frame matches the expected format ## For example, it should have columns: log2FoldChange, padj, etc. #res <- as.data.frame(res) ## Remove rows with NA in log2FoldChange (if needed) #res <- res[!is.na(res$log2FoldChange),] # Replace padj = 0 with a small value res$padj[res$padj == 0] <- 1e-12 #library(EnhancedVolcano) # Assuming res is already sorted and processed png("Mac.AB_vs_Mac.WT19606.png", width=1200, height=1200) #max.overlaps = 10 EnhancedVolcano(res, lab = rownames(res), x = 'log2FoldChange', y = 'padj', pCutoff = 1e-2, FCcutoff = 2, title = '', subtitleLabSize = 18, pointSize = 3.0, labSize = 5.0, colAlpha = 1, legendIconSize = 4.0, drawConnectors = TRUE, widthConnectors = 0.5, colConnectors = 'black', subtitle = expression("Mac.AB versus Mac.WT19606")) dev.off() # ---- Mac.IJ_vs_Mac.WT19606 ---- res <- read.csv("Mac.IJ_vs_Mac.WT19606-all.csv") # Replace empty GeneName with modified GeneID res$GeneName <- ifelse( res$GeneName == "" | is.na(res$GeneName), gsub("gene-", "", res$GeneID), res$GeneName ) duplicated_genes <- res[duplicated(res$GeneName), "GeneName"] res <- res %>% group_by(GeneName) %>% slice_min(padj, with_ties = FALSE) %>% ungroup() res <- as.data.frame(res) # Sort res first by padj (ascending) and then by log2FoldChange (descending) res <- res[order(res$padj, -res$log2FoldChange), ] # Assuming res is your dataframe and already processed # Filter up-regulated genes: log2FoldChange > 2 and padj < 1e-2 up_regulated <- res[res$log2FoldChange > 2 & res$padj < 1e-2, ] # Filter down-regulated genes: log2FoldChange < -2 and padj < 1e-2 down_regulated <- res[res$log2FoldChange < -2 & res$padj < 1e-2, ] # Create a new workbook wb <- createWorkbook() # Add the complete dataset as the first sheet addWorksheet(wb, "Complete_Data") writeData(wb, "Complete_Data", res) # Add the up-regulated genes as the second sheet addWorksheet(wb, "Up_Regulated") writeData(wb, "Up_Regulated", up_regulated) # Add the down-regulated genes as the third sheet addWorksheet(wb, "Down_Regulated") writeData(wb, "Down_Regulated", down_regulated) # Save the workbook to a file saveWorkbook(wb, "Gene_Expression_Mac.IJ_vs_Mac.WT19606.xlsx", overwrite = TRUE) # Set the 'GeneName' column as row.names rownames(res) <- res$GeneName # Drop the 'GeneName' column since it's now the row names res$GeneName <- NULL head(res) ## Ensure the data frame matches the expected format ## For example, it should have columns: log2FoldChange, padj, etc. #res <- as.data.frame(res) ## Remove rows with NA in log2FoldChange (if needed) #res <- res[!is.na(res$log2FoldChange),] # Replace padj = 0 with a small value res$padj[res$padj == 0] <- 1e-12 #library(EnhancedVolcano) # Assuming res is already sorted and processed png("Mac.IJ_vs_Mac.WT19606.png", width=1200, height=1200) #max.overlaps = 10 EnhancedVolcano(res, lab = rownames(res), x = 'log2FoldChange', y = 'padj', pCutoff = 1e-2, FCcutoff = 2, title = '', subtitleLabSize = 18, pointSize = 3.0, labSize = 5.0, colAlpha = 1, legendIconSize = 4.0, drawConnectors = TRUE, widthConnectors = 0.5, colConnectors = 'black', subtitle = expression("Mac.IJ versus Mac.WT19606")) dev.off() # ---- Mac.W1_vs_Mac.WT19606 ---- res <- read.csv("Mac.W1_vs_Mac.WT19606-all.csv") # Replace empty GeneName with modified GeneID res$GeneName <- ifelse( res$GeneName == "" | is.na(res$GeneName), gsub("gene-", "", res$GeneID), res$GeneName ) duplicated_genes <- res[duplicated(res$GeneName), "GeneName"] res <- res %>% group_by(GeneName) %>% slice_min(padj, with_ties = FALSE) %>% ungroup() res <- as.data.frame(res) # Sort res first by padj (ascending) and then by log2FoldChange (descending) res <- res[order(res$padj, -res$log2FoldChange), ] # Assuming res is your dataframe and already processed # Filter up-regulated genes: log2FoldChange > 2 and padj < 1e-2 up_regulated <- res[res$log2FoldChange > 2 & res$padj < 1e-2, ] # Filter down-regulated genes: log2FoldChange < -2 and padj < 1e-2 down_regulated <- res[res$log2FoldChange < -2 & res$padj < 1e-2, ] # Create a new workbook wb <- createWorkbook() # Add the complete dataset as the first sheet addWorksheet(wb, "Complete_Data") writeData(wb, "Complete_Data", res) # Add the up-regulated genes as the second sheet addWorksheet(wb, "Up_Regulated") writeData(wb, "Up_Regulated", up_regulated) # Add the down-regulated genes as the third sheet addWorksheet(wb, "Down_Regulated") writeData(wb, "Down_Regulated", down_regulated) # Save the workbook to a file saveWorkbook(wb, "Gene_Expression_Mac.W1_vs_Mac.WT19606.xlsx", overwrite = TRUE) # Set the 'GeneName' column as row.names rownames(res) <- res$GeneName # Drop the 'GeneName' column since it's now the row names res$GeneName <- NULL head(res) ## Ensure the data frame matches the expected format ## For example, it should have columns: log2FoldChange, padj, etc. #res <- as.data.frame(res) ## Remove rows with NA in log2FoldChange (if needed) #res <- res[!is.na(res$log2FoldChange),] # Replace padj = 0 with a small value res$padj[res$padj == 0] <- 1e-12 #library(EnhancedVolcano) # Assuming res is already sorted and processed png("Mac.W1_vs_Mac.WT19606.png", width=1200, height=1200) #max.overlaps = 10 EnhancedVolcano(res, lab = rownames(res), x = 'log2FoldChange', y = 'padj', pCutoff = 1e-2, FCcutoff = 2, title = '', subtitleLabSize = 18, pointSize = 3.0, labSize = 5.0, colAlpha = 1, legendIconSize = 4.0, drawConnectors = TRUE, widthConnectors = 0.5, colConnectors = 'black', subtitle = expression("Mac.W1 versus Mac.WT19606")) dev.off() # ---- Mac.Y1_vs_Mac.WT19606 ---- res <- read.csv("Mac.Y1_vs_Mac.WT19606-all.csv") # Replace empty GeneName with modified GeneID res$GeneName <- ifelse( res$GeneName == "" | is.na(res$GeneName), gsub("gene-", "", res$GeneID), res$GeneName ) duplicated_genes <- res[duplicated(res$GeneName), "GeneName"] res <- res %>% group_by(GeneName) %>% slice_min(padj, with_ties = FALSE) %>% ungroup() res <- as.data.frame(res) # Sort res first by padj (ascending) and then by log2FoldChange (descending) res <- res[order(res$padj, -res$log2FoldChange), ] # Assuming res is your dataframe and already processed # Filter up-regulated genes: log2FoldChange > 2 and padj < 1e-2 up_regulated <- res[res$log2FoldChange > 2 & res$padj < 1e-2, ] # Filter down-regulated genes: log2FoldChange < -2 and padj < 1e-2 down_regulated <- res[res$log2FoldChange < -2 & res$padj < 1e-2, ] # Create a new workbook wb <- createWorkbook() # Add the complete dataset as the first sheet addWorksheet(wb, "Complete_Data") writeData(wb, "Complete_Data", res) # Add the up-regulated genes as the second sheet addWorksheet(wb, "Up_Regulated") writeData(wb, "Up_Regulated", up_regulated) # Add the down-regulated genes as the third sheet addWorksheet(wb, "Down_Regulated") writeData(wb, "Down_Regulated", down_regulated) # Save the workbook to a file saveWorkbook(wb, "Gene_Expression_Mac.Y1_vs_Mac.WT19606.xlsx", overwrite = TRUE) # Set the 'GeneName' column as row.names rownames(res) <- res$GeneName # Drop the 'GeneName' column since it's now the row names res$GeneName <- NULL head(res) ## Ensure the data frame matches the expected format ## For example, it should have columns: log2FoldChange, padj, etc. #res <- as.data.frame(res) ## Remove rows with NA in log2FoldChange (if needed) #res <- res[!is.na(res$log2FoldChange),] # Replace padj = 0 with a small value res$padj[res$padj == 0] <- 1e-12 #library(EnhancedVolcano) # Assuming res is already sorted and processed png("Mac.Y1_vs_Mac.WT19606.png", width=1200, height=1200) #max.overlaps = 10 EnhancedVolcano(res, lab = rownames(res), x = 'log2FoldChange', y = 'padj', pCutoff = 1e-2, FCcutoff = 2, title = '', subtitleLabSize = 18, pointSize = 3.0, labSize = 5.0, colAlpha = 1, legendIconSize = 4.0, drawConnectors = TRUE, widthConnectors = 0.5, colConnectors = 'black', subtitle = expression("Mac.Y1 versus Mac.WT19606")) dev.off() #TODO: annotate the Gene_Expression_xxx_vs_yyy.xlsx -
Clustering the genes and draw heatmap
#http://xgenes.com/article/article-content/150/draw-venn-diagrams-using-matplotlib/ #http://xgenes.com/article/article-content/276/go-terms-for-s-epidermidis/ # save the Up-regulated and Down-regulated genes into -up.id and -down.id for i in Mac_vs_LB LB.AB_vs_LB.WT19606 LB.IJ_vs_LB.WT19606 LB.W1_vs_LB.WT19606 LB.Y1_vs_LB.WT19606 Mac.AB_vs_Mac.WT19606 Mac.IJ_vs_Mac.WT19606 Mac.W1_vs_Mac.WT19606 Mac.Y1_vs_Mac.WT19606; do echo "cut -d',' -f1-1 ${i}-up.txt > ${i}-up.id"; echo "cut -d',' -f1-1 ${i}-down.txt > ${i}-down.id"; done #5 LB.AB_vs_LB.WT19606-down.id #20 LB.AB_vs_LB.WT19606-up.id #64 LB.IJ_vs_LB.WT19606-down.id #69 LB.IJ_vs_LB.WT19606-up.id #23 LB.W1_vs_LB.WT19606-down.id #97 LB.W1_vs_LB.WT19606-up.id #9 LB.Y1_vs_LB.WT19606-down.id #20 LB.Y1_vs_LB.WT19606-up.id #20 Mac.AB_vs_Mac.WT19606-down.id #29 Mac.AB_vs_Mac.WT19606-up.id #65 Mac.IJ_vs_Mac.WT19606-down.id #197 Mac.IJ_vs_Mac.WT19606-up.id #359 Mac_vs_LB-down.id #308 Mac_vs_LB-up.id #290 Mac.W1_vs_Mac.WT19606-down.id #343 Mac.W1_vs_Mac.WT19606-up.id #75 Mac.Y1_vs_Mac.WT19606-down.id #0 Mac.Y1_vs_Mac.WT19606.png-down.id #0 Mac.Y1_vs_Mac.WT19606.png-up.id #68 Mac.Y1_vs_Mac.WT19606-up.id #2061 total cat *.id | sort -u > ids #Delete "GeneName" #add Gene_Id in the first line, delete the "" #Note that using GeneID as index, rather than GeneName, since .txt contains only GeneID. GOI <- read.csv("ids")$Gene_Id #1329 RNASeq.NoCellLine <- assay(rld) #install.packages("gplots") library("gplots") #clustering methods: "ward.D", "ward.D2", "single", "complete", "average" (= UPGMA), "mcquitty" (= WPGMA), "median" (= WPGMC) or "centroid" (= UPGMC). pearson or spearman datamat = RNASeq.NoCellLine[GOI, ] #datamat = RNASeq.NoCellLine write.csv(as.data.frame(datamat), file ="DEGs_heatmap_expression_data.txt") constant_rows <- apply(datamat, 1, function(row) var(row) == 0) if(any(constant_rows)) { cat("Removing", sum(constant_rows), "constant rows.\n") datamat <- datamat[!constant_rows, ] } hr <- hclust(as.dist(1-cor(t(datamat), method="pearson")), method="complete") hc <- hclust(as.dist(1-cor(datamat, method="spearman")), method="complete") mycl = cutree(hr, h=max(hr$height)/1.15) mycol = c("YELLOW", "BLUE", "ORANGE", "MAGENTA", "CYAN", "RED", "GREEN", "MAROON", "LIGHTBLUE", "PINK", "MAGENTA", "LIGHTCYAN", "LIGHTRED", "LIGHTGREEN"); mycol = mycol[as.vector(mycl)] #png("DEGs_heatmap.png", width=900, height=800) #cex.lab=10, labRow="", png("DEGs_heatmap.png", width=1200, height=1000) heatmap.2(as.matrix(datamat),Rowv=as.dendrogram(hr),Colv = NA, dendrogram = 'row',labRow="", scale='row',trace='none',col=bluered(75), cexCol=1.8, RowSideColors = mycol, margins=c(10,2), cexRow=1.5, srtCol=30, lhei = c(1, 8), lwid=c(2, 8)) #rownames(datamat) #heatmap.2(datamat, Rowv=as.dendrogram(hr), col=bluered(75), scale="row", RowSideColors=mycol, trace="none", margin=c(5,5), sepwidth=c(0,0), dendrogram = 'row', Colv = 'false', density.info='none', labRow="", srtCol=30, lhei=c(0.1,2)) dev.off() #### cluster members ##### write.csv(names(subset(mycl, mycl == '1')),file='cluster1_YELLOW.txt') write.csv(names(subset(mycl, mycl == '2')),file='cluster2_DARKBLUE.txt') write.csv(names(subset(mycl, mycl == '3')),file='cluster3_DARKORANGE.txt') write.csv(names(subset(mycl, mycl == '4')),file='cluster4_DARKMAGENTA.txt') write.csv(names(subset(mycl, mycl == '5')),file='cluster5_DARKCYAN.txt') #~/Tools/csv2xls-0.4/csv_to_xls.py cluster*.txt -d',' -o DEGs_heatmap_cluster_members.xls #~/Tools/csv2xls-0.4/csv_to_xls.py DEGs_heatmap_expression_data.txt -d',' -o DEGs_heatmap_expression_data.xls; #### (NOT_WORKING) cluster members (adding annotations, note that it does not work for the bacteria, since it is not model-speices and we cannot use mart=ensembl) ##### subset_1<-names(subset(mycl, mycl == '1')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_1, ]) #2575 subset_2<-names(subset(mycl, mycl == '2')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_2, ]) #1855 subset_3<-names(subset(mycl, mycl == '3')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_3, ]) #217 subset_4<-names(subset(mycl, mycl == '4')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_4, ]) # subset_5<-names(subset(mycl, mycl == '5')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_5, ]) # # Initialize an empty data frame for the annotated data annotated_data <- data.frame() # Determine total number of genes total_genes <- length(rownames(data)) # Loop through each gene to annotate for (i in 1:total_genes) { gene <- rownames(data)[i] result <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = gene, mart = ensembl) # If multiple rows are returned, take the first one if (nrow(result) > 1) { result <- result[1, ] } # Check if the result is empty if (nrow(result) == 0) { result <- data.frame(ensembl_gene_id = gene, external_gene_name = NA, gene_biotype = NA, entrezgene_id = NA, chromosome_name = NA, start_position = NA, end_position = NA, strand = NA, description = NA) } # Transpose expression values expression_values <- t(data.frame(t(data[gene, ]))) colnames(expression_values) <- colnames(data) # Combine gene information and expression data combined_result <- cbind(result, expression_values) # Append to the final dataframe annotated_data <- rbind(annotated_data, combined_result) # Print progress every 100 genes if (i %% 100 == 0) { cat(sprintf("Processed gene %d out of %d\n", i, total_genes)) } } # Save the annotated data to a new CSV file write.csv(annotated_data, "cluster1_YELLOW.csv", row.names=FALSE) write.csv(annotated_data, "cluster2_DARKBLUE.csv", row.names=FALSE) write.csv(annotated_data, "cluster3_DARKORANGE.csv", row.names=FALSE) write.csv(annotated_data, "cluster4_DARKMAGENTA.csv", row.names=FALSE) write.csv(annotated_data, "cluster5_DARKCYAN.csv", row.names=FALSE) #~/Tools/csv2xls-0.4/csv_to_xls.py cluster*.csv -d',' -o DEGs_heatmap_clusters.xls
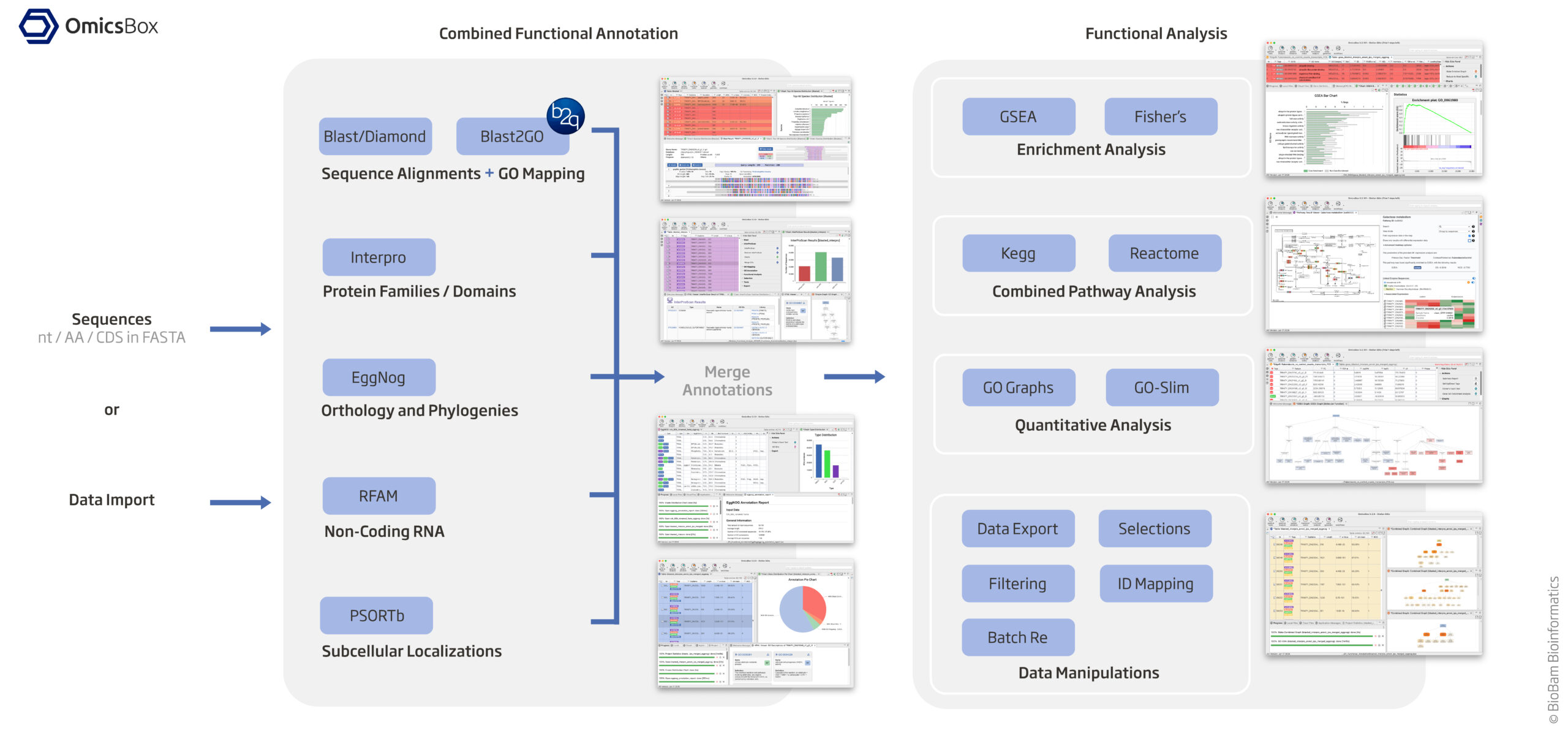
KEGG and GO annotations in non-model organisms
https://www.biobam.com/functional-analysis/
-
Assign KEGG and GO Terms (see diagram above)
Since your organism is non-model, standard R databases (org.Hs.eg.db, etc.) won’t work. You’ll need to manually retrieve KEGG and GO annotations.
Option 1 (KEGG Terms): EggNog based on orthology and phylogenies
EggNOG-mapper assigns both KEGG Orthology (KO) IDs and GO terms. Install EggNOG-mapper: mamba create -n eggnog_env python=3.8 eggnog-mapper -c conda-forge -c bioconda #eggnog-mapper_2.1.12 mamba activate eggnog_env Run annotation: #diamond makedb --in eggnog6.prots.faa -d eggnog_proteins.dmnd mkdir /home/jhuang/mambaforge/envs/eggnog_env/lib/python3.8/site-packages/data/ download_eggnog_data.py --dbname eggnog.db -y --data_dir /home/jhuang/mambaforge/envs/eggnog_env/lib/python3.8/site-packages/data/ #NOT_WORKING: emapper.py -i CP059040_gene.fasta -o eggnog_dmnd_out --cpu 60 -m diamond[hmmer,mmseqs] --dmnd_db /home/jhuang/REFs/eggnog_data/data/eggnog_proteins.dmnd python ~/Scripts/update_fasta_header.py CP059040_protein_.fasta CP059040_protein.fasta emapper.py -i CP059040_protein.fasta -o eggnog_out --cpu 60 --resume #----> result annotations.tsv: Contains KEGG, GO, and other functional annotations. #----> 470.IX87_14445: * 470 likely refers to the organism or strain (e.g., Acinetobacter baumannii ATCC 19606 or another related strain). * IX87_14445 would refer to a specific gene or protein within that genome. Extract KEGG KO IDs from annotations.emapper.annotations.Option 2 (GO Terms from 'Blast2GO 5 Basic', saved in blast2go_annot.annot): Using Blast/Diamond + Blast2GO_GUI based on sequence alignment + GO mapping
- 'Load protein sequences' (Tags: NONE, generated columns: Nr, SeqName) -->
- Buttons 'blast' (Tags: BLASTED, generated columns: Description, Length, #Hits, e-Value, sim mean),
- Button 'mapping' (Tags: MAPPED, generated columns: #GO, GO IDs, GO Names), "Mapping finished - Please proceed now to annotation."
- Button 'annot' (Tags: ANNOTATED, generated columns: Enzyme Codes, Enzyme Names), "Annotation finished." * Used parameter 'Annotation CutOff': The Blast2GO Annotation Rule seeks to find the most specific GO annotations with a certain level of reliability. An annotation score is calculated for each candidate GO which is composed by the sequence similarity of the Blast Hit, the evidence code of the source GO and the position of the particular GO in the Gene Ontology hierarchy. This annotation score cutoff select the most specific GO term for a given GO branch which lies above this value. * Used parameter 'GO Weight' is a value which is added to Annotation Score of a more general/abstract Gene Ontology term for each of its more specific, original source GO terms. In this case, more general GO terms which summarise many original source terms (those ones directly associated to the Blast Hits) will have a higher Annotation Score.
or blast2go_cli_v1.5.1 (NOT_USED)
#https://help.biobam.com/space/BCD/2250407989/Installation #see ~/Scripts/blast2go_pipeline.shOption 3 (GO Terms from 'Blast2GO 5 Basic', saved in blast2go_annot.annot2): Interpro based protein families / domains --> Button interpro * Button 'interpro' (Tags: INTERPRO, generated columns: InterPro IDs, InterPro GO IDs, InterPro GO Names) --> "InterProScan Finished - You can now merge the obtained GO Annotations."
MERGE the results of InterPro GO IDs (Option 3) to GO IDs (Option 2) and generate final GO IDs * Button 'interpro'/'Merge InterProScan GOs to Annotation' --> "Merge (add and validate) all GO terms retrieved via InterProScan to the already existing GO annotation." --> "Finished merging GO terms from InterPro with annotations. Maybe you want to run ANNEX (Annotation Augmentation)." #* Button 'annot'/'ANNEX' --> "ANNEX finished. Maybe you want to do the next step: Enzyme Code Mapping."
#-- before merging (blast2go_annot.annot) -- #H0N29_18790 GO:0004842 ankyrin repeat domain-containing protein #H0N29_18790 GO:0085020 #-- after merging (blast2go_annot.annot2) --> #H0N29_18790 GO:0031436 ankyrin repeat domain-containing protein #H0N29_18790 GO:0070531 #H0N29_18790 GO:0004842 #H0N29_18790 GO:0005515 #H0N29_18790 GO:0085020Option 4 (NOT_USED): RFAM for non-colding RNA
Option 5 (NOT_USED): PSORTb for subcellular localizations
Option 6 (NOT_USED): KAAS (KEGG Automatic Annotation Server)
- Go to KAAS
- Upload your FASTA file.
- Select an appropriate gene set.
- Download the KO assignments.
-
Find the Closest KEGG Organism Code (NOT_USED)
Since your species isn't directly in KEGG, use a closely related organism.
- Check available KEGG organisms:
library(clusterProfiler) library(KEGGREST) kegg_organisms <- keggList("organism") Pick the closest relative (e.g., zebrafish "dre" for fish, Arabidopsis "ath" for plants). # Search for Acinetobacter in the list grep("Acinetobacter", kegg_organisms, ignore.case = TRUE, value = TRUE) # Gammaproteobacteria #Extract KO IDs from the eggnog results for "Acinetobacter baumannii strain ATCC 19606"
- Check available KEGG organisms:
-
Find the Closest KEGG Organism for a Non-Model Species
If your organism is not in KEGG, search for the closest relative:
grep("fish", kegg_organisms, ignore.case = TRUE, value = TRUE) # Example searchFor KEGG pathway enrichment in non-model species, use "ko" instead of a species code (the code has been intergrated in the point 4):
kegg_enrich <- enrichKEGG(gene = gene_list, organism = "ko") # "ko" = KEGG Orthology -
Perform KEGG and GO Enrichment in R (under dir ~/DATA/ata_Tam_RNAseq_2025_LB_vs_Mac_ATCC19606/results/star_salmon/degenes)
#BiocManager::install("GO.db") #BiocManager::install("AnnotationDbi") # Load required libraries library(openxlsx) # For Excel file handling library(dplyr) # For data manipulation library(tidyr) library(stringr) library(clusterProfiler) # For KEGG and GO enrichment analysis #library(org.Hs.eg.db) # Replace with appropriate organism database library(GO.db) library(AnnotationDbi) setwd("~/DATA/Data_Tam_RNAseq_2025_LB_vs_Mac_ATCC19606/results/star_salmon/degenes") # PREPARING go_terms and ec_terms: annot_* file: cut -f1-2 -d$'\t' blast2go_annot.annot2 > blast2go_annot.annot2_ # Step 1: Load the blast2go annotation file with a check for missing columns annot_df <- read.table("/home/jhuang/b2gWorkspace_Tam_RNAseq_2024/blast2go_annot.annot2_", header = FALSE, sep = "\t", stringsAsFactors = FALSE, fill = TRUE) # If the structure is inconsistent, we can make sure there are exactly 3 columns: colnames(annot_df) <- c("GeneID", "Term") # Step 2: Filter and aggregate GO and EC terms as before go_terms <- annot_df %>% filter(grepl("^GO:", Term)) %>% group_by(GeneID) %>% summarize(GOs = paste(Term, collapse = ","), .groups = "drop") ec_terms <- annot_df %>% filter(grepl("^EC:", Term)) %>% group_by(GeneID) %>% summarize(EC = paste(Term, collapse = ","), .groups = "drop") # Load the results #res <- read.csv("Mac_vs_LB-all.csv") #up307, down358 #res <- read.csv("LB.AB_vs_LB.WT19606-all.csv") #up307, down358 #res <- read.csv("LB.IJ_vs_LB.WT19606-all.csv") #up307, down358 #res <- read.csv("LB.W1_vs_LB.WT19606-all.csv") #up307, down358 #res <- read.csv("LB.Y1_vs_LB.WT19606-all.csv") #up307, down358 #res <- read.csv("Mac.AB_vs_Mac.WT19606-all.csv") #up307, down358 #res <- read.csv("Mac.IJ_vs_Mac.WT19606-all.csv") #up307, down358 #res <- read.csv("Mac.W1_vs_Mac.WT19606-all.csv") #up307, down358 res <- read.csv("Mac.Y1_vs_Mac.WT19606-all.csv") #up307, down358 # Replace empty GeneName with modified GeneID res$GeneName <- ifelse( res$GeneName == "" | is.na(res$GeneName), gsub("gene-", "", res$GeneID), res$GeneName ) # Remove duplicated genes by selecting the gene with the smallest padj duplicated_genes <- res[duplicated(res$GeneName), "GeneName"] res <- res %>% group_by(GeneName) %>% slice_min(padj, with_ties = FALSE) %>% ungroup() res <- as.data.frame(res) # Sort res first by padj (ascending) and then by log2FoldChange (descending) res <- res[order(res$padj, -res$log2FoldChange), ] # Read eggnog annotations eggnog_data <- read.delim("~/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine_ATCC19606/eggnog_out.emapper.annotations.txt", header = TRUE, sep = "\t") # Remove the "gene-" prefix from GeneID in res to match eggnog 'query' format res$GeneID <- gsub("gene-", "", res$GeneID) # Merge eggnog data with res based on GeneID res <- res %>% left_join(eggnog_data, by = c("GeneID" = "query")) # Merge with the res dataframe # Perform the left joins and rename columns res_updated <- res %>% left_join(go_terms, by = "GeneID") %>% left_join(ec_terms, by = "GeneID") %>% dplyr::select(-EC.x, -GOs.x) %>% dplyr::rename(EC = EC.y, GOs = GOs.y) # Filter up-regulated genes up_regulated <- res_updated[res_updated$log2FoldChange > 2 & res_updated$padj < 0.01, ] # Filter down-regulated genes down_regulated <- res_updated[res_updated$log2FoldChange < -2 & res_updated$padj < 0.01, ] # Create a new workbook wb <- createWorkbook() # Add the complete dataset as the first sheet (with annotations) addWorksheet(wb, "Complete_Data") writeData(wb, "Complete_Data", res_updated) # Add the up-regulated genes as the second sheet (with annotations) addWorksheet(wb, "Up_Regulated") writeData(wb, "Up_Regulated", up_regulated) # Add the down-regulated genes as the third sheet (with annotations) addWorksheet(wb, "Down_Regulated") writeData(wb, "Down_Regulated", down_regulated) # Save the workbook to a file saveWorkbook(wb, "Gene_Expression_with_Annotations_Urine_vs_MHB.xlsx", overwrite = TRUE) # Set GeneName as row names after the join rownames(res_updated) <- res_updated$GeneName res_updated <- res_updated %>% dplyr::select(-GeneName) ## Set the 'GeneName' column as row.names #rownames(res_updated) <- res_updated$GeneName ## Drop the 'GeneName' column since it's now the row names #res_updated$GeneName <- NULL # -- BREAK_1 -- # ---- Perform KEGG enrichment analysis (up_regulated) ---- gene_list_kegg_up <- up_regulated$KEGG_ko gene_list_kegg_up <- gsub("ko:", "", gene_list_kegg_up) kegg_enrichment_up <- enrichKEGG(gene = gene_list_kegg_up, organism = 'ko') # -- convert the GeneID (Kxxxxxx) to the true GeneID -- # Step 0: Create KEGG to GeneID mapping kegg_to_geneid_up <- up_regulated %>% dplyr::select(KEGG_ko, GeneID) %>% filter(!is.na(KEGG_ko)) %>% # Remove missing KEGG KO entries mutate(KEGG_ko = str_remove(KEGG_ko, "ko:")) # Remove 'ko:' prefix if present # Step 1: Clean KEGG_ko values (separate multiple KEGG IDs) kegg_to_geneid_clean <- kegg_to_geneid_up %>% mutate(KEGG_ko = str_remove_all(KEGG_ko, "ko:")) %>% # Remove 'ko:' prefixes separate_rows(KEGG_ko, sep = ",") %>% # Ensure each KEGG ID is on its own row filter(KEGG_ko != "-") %>% # Remove invalid KEGG IDs ("-") distinct() # Remove any duplicate mappings # Step 2.1: Expand geneID column in kegg_enrichment_up expanded_kegg <- kegg_enrichment_up %>% as.data.frame() %>% separate_rows(geneID, sep = "/") %>% # Split multiple KEGG IDs (Kxxxxx) left_join(kegg_to_geneid_clean, by = c("geneID" = "KEGG_ko"), relationship = "many-to-many") %>% # Explicitly handle many-to-many distinct() %>% # Remove duplicate matches group_by(ID) %>% summarise(across(everything(), ~ paste(unique(na.omit(.)), collapse = "/")), .groups = "drop") # Re-collapse results #dplyr::glimpse(expanded_kegg) # Step 3.1: Replace geneID column in the original dataframe kegg_enrichment_up_df <- as.data.frame(kegg_enrichment_up) # Remove old geneID column and merge new one kegg_enrichment_up_df <- kegg_enrichment_up_df %>% dplyr::select(-geneID) %>% # Remove old geneID column left_join(expanded_kegg %>% dplyr::select(ID, GeneID), by = "ID") %>% # Merge new GeneID column dplyr::rename(geneID = GeneID) # Rename column back to geneID # ---- Perform KEGG enrichment analysis (down_regulated) ---- # Step 1: Extract KEGG KO terms from down-regulated genes gene_list_kegg_down <- down_regulated$KEGG_ko gene_list_kegg_down <- gsub("ko:", "", gene_list_kegg_down) # Step 2: Perform KEGG enrichment analysis kegg_enrichment_down <- enrichKEGG(gene = gene_list_kegg_down, organism = 'ko') # --- Convert KEGG gene IDs (Kxxxxxx) to actual GeneIDs --- # Step 3: Create KEGG to GeneID mapping from down_regulated dataset kegg_to_geneid_down <- down_regulated %>% dplyr::select(KEGG_ko, GeneID) %>% filter(!is.na(KEGG_ko)) %>% # Remove missing KEGG KO entries mutate(KEGG_ko = str_remove(KEGG_ko, "ko:")) # Remove 'ko:' prefix if present # -- BREAK_2 -- # Step 4: Clean KEGG_ko values (handle multiple KEGG IDs) kegg_to_geneid_down_clean <- kegg_to_geneid_down %>% mutate(KEGG_ko = str_remove_all(KEGG_ko, "ko:")) %>% # Remove 'ko:' prefixes separate_rows(KEGG_ko, sep = ",") %>% # Ensure each KEGG ID is on its own row filter(KEGG_ko != "-") %>% # Remove invalid KEGG IDs ("-") distinct() # Remove duplicate mappings # Step 5: Expand geneID column in kegg_enrichment_down expanded_kegg_down <- kegg_enrichment_down %>% as.data.frame() %>% separate_rows(geneID, sep = "/") %>% # Split multiple KEGG IDs (Kxxxxx) left_join(kegg_to_geneid_down_clean, by = c("geneID" = "KEGG_ko"), relationship = "many-to-many") %>% # Handle many-to-many mappings distinct() %>% # Remove duplicate matches group_by(ID) %>% summarise(across(everything(), ~ paste(unique(na.omit(.)), collapse = "/")), .groups = "drop") # Re-collapse results # Step 6: Replace geneID column in the original kegg_enrichment_down dataframe kegg_enrichment_down_df <- as.data.frame(kegg_enrichment_down) %>% dplyr::select(-geneID) %>% # Remove old geneID column left_join(expanded_kegg_down %>% dplyr::select(ID, GeneID), by = "ID") %>% # Merge new GeneID column dplyr::rename(geneID = GeneID) # Rename column back to geneID # View the updated dataframe head(kegg_enrichment_down_df) # Create a new workbook wb <- createWorkbook() # Save enrichment results to the workbook addWorksheet(wb, "KEGG_Enrichment_Up") writeData(wb, "KEGG_Enrichment_Up", as.data.frame(kegg_enrichment_up_df)) # Save enrichment results to the workbook addWorksheet(wb, "KEGG_Enrichment_Down") writeData(wb, "KEGG_Enrichment_Down", as.data.frame(kegg_enrichment_down_df)) # Define gene list (up-regulated genes) gene_list_go_up <- up_regulated$GeneID # Extract the 149 up-regulated genes gene_list_go_down <- down_regulated$GeneID # Extract the 65 down-regulated genes # Define background gene set (all genes in res) background_genes <- res_updated$GeneID # Extract the 3646 background genes # Prepare GO annotation data from res go_annotation <- res_updated[, c("GOs","GeneID")] # Extract relevant columns go_annotation <- go_annotation %>% tidyr::separate_rows(GOs, sep = ",") # Split multiple GO terms into separate rows # -- BREAK_3 -- go_enrichment_up <- enricher( gene = gene_list_go_up, # Up-regulated genes TERM2GENE = go_annotation, # Custom GO annotation pvalueCutoff = 0.05, # Significance threshold pAdjustMethod = "BH", universe = background_genes # Define the background gene set ) go_enrichment_up <- as.data.frame(go_enrichment_up) go_enrichment_down <- enricher( gene = gene_list_go_down, # Up-regulated genes TERM2GENE = go_annotation, # Custom GO annotation pvalueCutoff = 0.05, # Significance threshold pAdjustMethod = "BH", universe = background_genes # Define the background gene set ) go_enrichment_down <- as.data.frame(go_enrichment_down) ## Remove the 'p.adjust' column since no adjusted methods have been applied --> In this version we have used pvalue filtering (see above)! #go_enrichment_up <- go_enrichment_up[, !names(go_enrichment_up) %in% "p.adjust"] # Update the Description column with the term descriptions go_enrichment_up$Description <- sapply(go_enrichment_up$ID, function(go_id) { # Using select to get the term description term <- tryCatch({ AnnotationDbi::select(GO.db, keys = go_id, columns = "TERM", keytype = "GOID") }, error = function(e) { message(paste("Error for GO term:", go_id)) # Print which GO ID caused the error return(data.frame(TERM = NA)) # In case of error, return NA }) if (nrow(term) > 0) { return(term$TERM) } else { return(NA) # If no description found, return NA } }) ## Print the updated data frame #print(go_enrichment_up) ## Remove the 'p.adjust' column since no adjusted methods have been applied --> In this version we have used pvalue filtering (see above)! #go_enrichment_down <- go_enrichment_down[, !names(go_enrichment_down) %in% "p.adjust"] # Update the Description column with the term descriptions go_enrichment_down$Description <- sapply(go_enrichment_down$ID, function(go_id) { # Using select to get the term description term <- tryCatch({ AnnotationDbi::select(GO.db, keys = go_id, columns = "TERM", keytype = "GOID") }, error = function(e) { message(paste("Error for GO term:", go_id)) # Print which GO ID caused the error return(data.frame(TERM = NA)) # In case of error, return NA }) if (nrow(term) > 0) { return(term$TERM) } else { return(NA) # If no description found, return NA } }) addWorksheet(wb, "GO_Enrichment_Up") writeData(wb, "GO_Enrichment_Up", as.data.frame(go_enrichment_up)) addWorksheet(wb, "GO_Enrichment_Down") writeData(wb, "GO_Enrichment_Down", as.data.frame(go_enrichment_down)) # Save the workbook with enrichment results saveWorkbook(wb, "KEGG_and_GO_Enrichments_Urine_vs_MHB.xlsx", overwrite = TRUE) #Error for GO term: GO:0006807: replace "GO:0006807 obsolete nitrogen compound metabolic process" #obsolete nitrogen compound metabolic process #https://www.ebi.ac.uk/QuickGO/term/GO:0006807 #TODO: marked the color as yellow if the p.adjusted <= 0.05 in GO_enrichment! #mv KEGG_and_GO_Enrichments_Urine_vs_MHB.xlsx KEGG_and_GO_Enrichments_Mac_vs_LB.xlsx #Mac_vs_LB #LB.AB_vs_LB.WT19606 #LB.IJ_vs_LB.WT19606 #LB.W1_vs_LB.WT19606 #LB.Y1_vs_LB.WT19606 #Mac.AB_vs_Mac.WT19606 #Mac.IJ_vs_Mac.WT19606 #Mac.W1_vs_Mac.WT19606 #Mac.Y1_vs_Mac.WT19606 -
(DEBUG) Draw the Venn diagram to compare the total DEGs across AUM, Urine, and MHB, irrespective of up- or down-regulation.
library(openxlsx) # Function to read and clean gene ID files read_gene_ids <- function(file_path) { # Read the gene IDs from the file gene_ids <- readLines(file_path) # Remove any quotes and trim whitespaces gene_ids <- gsub('"', '', gene_ids) # Remove quotes gene_ids <- trimws(gene_ids) # Trim whitespaces # Remove empty entries or NAs gene_ids <- gene_ids[gene_ids != "" & !is.na(gene_ids)] return(gene_ids) } # Example list of LB files with both -up.id and -down.id for each condition lb_files_up <- c("LB.AB_vs_LB.WT19606-up.id", "LB.IJ_vs_LB.WT19606-up.id", "LB.W1_vs_LB.WT19606-up.id", "LB.Y1_vs_LB.WT19606-up.id") lb_files_down <- c("LB.AB_vs_LB.WT19606-down.id", "LB.IJ_vs_LB.WT19606-down.id", "LB.W1_vs_LB.WT19606-down.id", "LB.Y1_vs_LB.WT19606-down.id") # Combine both up and down files for each condition lb_files <- c(lb_files_up, lb_files_down) # Read gene IDs for each file in LB group #lb_degs <- setNames(lapply(lb_files, read_gene_ids), gsub("-(up|down).id", "", lb_files)) lb_degs <- setNames(lapply(lb_files, read_gene_ids), make.unique(gsub("-(up|down).id", "", lb_files))) lb_degs_ <- list() combined_set <- c(lb_degs[["LB.AB_vs_LB.WT19606"]], lb_degs[["LB.AB_vs_LB.WT19606.1"]]) #unique_combined_set <- unique(combined_set) lb_degs_$AB <- combined_set combined_set <- c(lb_degs[["LB.IJ_vs_LB.WT19606"]], lb_degs[["LB.IJ_vs_LB.WT19606.1"]]) lb_degs_$IJ <- combined_set combined_set <- c(lb_degs[["LB.W1_vs_LB.WT19606"]], lb_degs[["LB.W1_vs_LB.WT19606.1"]]) lb_degs_$W1 <- combined_set combined_set <- c(lb_degs[["LB.Y1_vs_LB.WT19606"]], lb_degs[["LB.Y1_vs_LB.WT19606.1"]]) lb_degs_$Y1 <- combined_set # Example list of Mac files with both -up.id and -down.id for each condition mac_files_up <- c("Mac.AB_vs_Mac.WT19606-up.id", "Mac.IJ_vs_Mac.WT19606-up.id", "Mac.W1_vs_Mac.WT19606-up.id", "Mac.Y1_vs_Mac.WT19606-up.id") mac_files_down <- c("Mac.AB_vs_Mac.WT19606-down.id", "Mac.IJ_vs_Mac.WT19606-down.id", "Mac.W1_vs_Mac.WT19606-down.id", "Mac.Y1_vs_Mac.WT19606-down.id") # Combine both up and down files for each condition in Mac group mac_files <- c(mac_files_up, mac_files_down) # Read gene IDs for each file in Mac group mac_degs <- setNames(lapply(mac_files, read_gene_ids), make.unique(gsub("-(up|down).id", "", mac_files))) mac_degs_ <- list() combined_set <- c(mac_degs[["Mac.AB_vs_Mac.WT19606"]], mac_degs[["Mac.AB_vs_Mac.WT19606.1"]]) mac_degs_$AB <- combined_set combined_set <- c(mac_degs[["Mac.IJ_vs_Mac.WT19606"]], mac_degs[["Mac.IJ_vs_Mac.WT19606.1"]]) mac_degs_$IJ <- combined_set combined_set <- c(mac_degs[["Mac.W1_vs_Mac.WT19606"]], mac_degs[["Mac.W1_vs_Mac.WT19606.1"]]) mac_degs_$W1 <- combined_set combined_set <- c(mac_degs[["Mac.Y1_vs_Mac.WT19606"]], mac_degs[["Mac.Y1_vs_Mac.WT19606.1"]]) mac_degs_$Y1 <- combined_set # Function to clean sheet names to ensure no sheet name exceeds 31 characters truncate_sheet_name <- function(names_list) { sapply(names_list, function(name) { if (nchar(name) > 31) { return(substr(name, 1, 31)) # Truncate sheet name to 31 characters } return(name) }) } # Assuming lb_degs_ is already a list of gene sets (LB.AB, LB.IJ, etc.) # Define intersections between different conditions for LB inter_lb_ab_ij <- intersect(lb_degs_$AB, lb_degs_$IJ) inter_lb_ab_w1 <- intersect(lb_degs_$AB, lb_degs_$W1) inter_lb_ab_y1 <- intersect(lb_degs_$AB, lb_degs_$Y1) inter_lb_ij_w1 <- intersect(lb_degs_$IJ, lb_degs_$W1) inter_lb_ij_y1 <- intersect(lb_degs_$IJ, lb_degs_$Y1) inter_lb_w1_y1 <- intersect(lb_degs_$W1, lb_degs_$Y1) # Define intersections between three conditions for LB inter_lb_ab_ij_w1 <- Reduce(intersect, list(lb_degs_$AB, lb_degs_$IJ, lb_degs_$W1)) inter_lb_ab_ij_y1 <- Reduce(intersect, list(lb_degs_$AB, lb_degs_$IJ, lb_degs_$Y1)) inter_lb_ab_w1_y1 <- Reduce(intersect, list(lb_degs_$AB, lb_degs_$W1, lb_degs_$Y1)) inter_lb_ij_w1_y1 <- Reduce(intersect, list(lb_degs_$IJ, lb_degs_$W1, lb_degs_$Y1)) # Define intersection between all four conditions for LB inter_lb_ab_ij_w1_y1 <- Reduce(intersect, list(lb_degs_$AB, lb_degs_$IJ, lb_degs_$W1, lb_degs_$Y1)) # Now remove the intersected genes from each original set for LB venn_list_lb <- list() # For LB.AB, remove genes that are also in other conditions venn_list_lb[["LB.AB_only"]] <- setdiff(lb_degs_$AB, union(inter_lb_ab_ij, union(inter_lb_ab_w1, inter_lb_ab_y1))) # For LB.IJ, remove genes that are also in other conditions venn_list_lb[["LB.IJ_only"]] <- setdiff(lb_degs_$IJ, union(inter_lb_ab_ij, union(inter_lb_ij_w1, inter_lb_ij_y1))) # For LB.W1, remove genes that are also in other conditions venn_list_lb[["LB.W1_only"]] <- setdiff(lb_degs_$W1, union(inter_lb_ab_w1, union(inter_lb_ij_w1, inter_lb_ab_w1_y1))) # For LB.Y1, remove genes that are also in other conditions venn_list_lb[["LB.Y1_only"]] <- setdiff(lb_degs_$Y1, union(inter_lb_ab_y1, union(inter_lb_ij_y1, inter_lb_ab_w1_y1))) # Add the intersections for LB (same as before) venn_list_lb[["LB.AB_AND_LB.IJ"]] <- inter_lb_ab_ij venn_list_lb[["LB.AB_AND_LB.W1"]] <- inter_lb_ab_w1 venn_list_lb[["LB.AB_AND_LB.Y1"]] <- inter_lb_ab_y1 venn_list_lb[["LB.IJ_AND_LB.W1"]] <- inter_lb_ij_w1 venn_list_lb[["LB.IJ_AND_LB.Y1"]] <- inter_lb_ij_y1 venn_list_lb[["LB.W1_AND_LB.Y1"]] <- inter_lb_w1_y1 # Define intersections between three conditions for LB venn_list_lb[["LB.AB_AND_LB.IJ_AND_LB.W1"]] <- inter_lb_ab_ij_w1 venn_list_lb[["LB.AB_AND_LB.IJ_AND_LB.Y1"]] <- inter_lb_ab_ij_y1 venn_list_lb[["LB.AB_AND_LB.W1_AND_LB.Y1"]] <- inter_lb_ab_w1_y1 venn_list_lb[["LB.IJ_AND_LB.W1_AND_LB.Y1"]] <- inter_lb_ij_w1_y1 # Define intersection between all four conditions for LB venn_list_lb[["LB.AB_AND_LB.IJ_AND_LB.W1_AND_LB.Y1"]] <- inter_lb_ab_ij_w1_y1 # Assuming mac_degs_ is already a list of gene sets (Mac.AB, Mac.IJ, etc.) # Define intersections between different conditions inter_mac_ab_ij <- intersect(mac_degs_$AB, mac_degs_$IJ) inter_mac_ab_w1 <- intersect(mac_degs_$AB, mac_degs_$W1) inter_mac_ab_y1 <- intersect(mac_degs_$AB, mac_degs_$Y1) inter_mac_ij_w1 <- intersect(mac_degs_$IJ, mac_degs_$W1) inter_mac_ij_y1 <- intersect(mac_degs_$IJ, mac_degs_$Y1) inter_mac_w1_y1 <- intersect(mac_degs_$W1, mac_degs_$Y1) # Define intersections between three conditions inter_mac_ab_ij_w1 <- Reduce(intersect, list(mac_degs_$AB, mac_degs_$IJ, mac_degs_$W1)) inter_mac_ab_ij_y1 <- Reduce(intersect, list(mac_degs_$AB, mac_degs_$IJ, mac_degs_$Y1)) inter_mac_ab_w1_y1 <- Reduce(intersect, list(mac_degs_$AB, mac_degs_$W1, mac_degs_$Y1)) inter_mac_ij_w1_y1 <- Reduce(intersect, list(mac_degs_$IJ, mac_degs_$W1, mac_degs_$Y1)) # Define intersection between all four conditions inter_mac_ab_ij_w1_y1 <- Reduce(intersect, list(mac_degs_$AB, mac_degs_$IJ, mac_degs_$W1, mac_degs_$Y1)) # Now remove the intersected genes from each original set venn_list_mac <- list() # For Mac.AB, remove genes that are also in other conditions venn_list_mac[["Mac.AB_only"]] <- setdiff(mac_degs_$AB, union(inter_mac_ab_ij, union(inter_mac_ab_w1, inter_mac_ab_y1))) # For Mac.IJ, remove genes that are also in other conditions venn_list_mac[["Mac.IJ_only"]] <- setdiff(mac_degs_$IJ, union(inter_mac_ab_ij, union(inter_mac_ij_w1, inter_mac_ij_y1))) # For Mac.W1, remove genes that are also in other conditions venn_list_mac[["Mac.W1_only"]] <- setdiff(mac_degs_$W1, union(inter_mac_ab_w1, union(inter_mac_ij_w1, inter_mac_ab_w1_y1))) # For Mac.Y1, remove genes that are also in other conditions venn_list_mac[["Mac.Y1_only"]] <- setdiff(mac_degs_$Y1, union(inter_mac_ab_y1, union(inter_mac_ij_y1, inter_mac_ab_w1_y1))) # Add the intersections (same as before) venn_list_mac[["Mac.AB_AND_Mac.IJ"]] <- inter_mac_ab_ij venn_list_mac[["Mac.AB_AND_Mac.W1"]] <- inter_mac_ab_w1 venn_list_mac[["Mac.AB_AND_Mac.Y1"]] <- inter_mac_ab_y1 venn_list_mac[["Mac.IJ_AND_Mac.W1"]] <- inter_mac_ij_w1 venn_list_mac[["Mac.IJ_AND_Mac.Y1"]] <- inter_mac_ij_y1 venn_list_mac[["Mac.W1_AND_Mac.Y1"]] <- inter_mac_w1_y1 # Define intersections between three conditions venn_list_mac[["Mac.AB_AND_Mac.IJ_AND_Mac.W1"]] <- inter_mac_ab_ij_w1 venn_list_mac[["Mac.AB_AND_Mac.IJ_AND_Mac.Y1"]] <- inter_mac_ab_ij_y1 venn_list_mac[["Mac.AB_AND_Mac.W1_AND_Mac.Y1"]] <- inter_mac_ab_w1_y1 venn_list_mac[["Mac.IJ_AND_Mac.W1_AND_Mac.Y1"]] <- inter_mac_ij_w1_y1 # Define intersection between all four conditions venn_list_mac[["Mac.AB_AND_Mac.IJ_AND_Mac.W1_AND_Mac.Y1"]] <- inter_mac_ab_ij_w1_y1 # Save the gene IDs to Excel for further inspection (optional) write.xlsx(lb_degs, file = "LB_DEGs.xlsx") write.xlsx(mac_degs, file = "Mac_DEGs.xlsx") # Clean sheet names and write the Venn intersection sets for LB and Mac groups into Excel files write.xlsx(venn_list_lb, file = "Venn_LB_Genes_Intersect.xlsx", sheetName = truncate_sheet_name(names(venn_list_lb)), rowNames = FALSE) write.xlsx(venn_list_mac, file = "Venn_Mac_Genes_Intersect.xlsx", sheetName = truncate_sheet_name(names(venn_list_mac)), rowNames = FALSE) # Venn Diagram for LB group venn1 <- ggvenn(lb_degs_, fill_color = c("skyblue", "tomato", "gold", "orchid"), stroke_size = 0.4, set_name_size = 5) ggsave("Venn_LB_Genes.png", plot = venn1, width = 7, height = 7, dpi = 300) # Venn Diagram for Mac group venn2 <- ggvenn(mac_degs_, fill_color = c("lightgreen", "slateblue", "plum", "orange"), stroke_size = 0.4, set_name_size = 5) ggsave("Venn_Mac_Genes.png", plot = venn2, width = 7, height = 7, dpi = 300) cat("✅ All Venn intersection sets exported to Excel successfully.\n")
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Kraken2 Installation and Usage Guide
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Should the inputs for GSVA be normalized or raw?
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- Risks of Rebooting into Rescue Mode
- 足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Processing RNAseq_2025_WT_vs_ΔIJ_on_ATCC19606
How to correlate RNA-seq Data with Mass Spectrometry Proteomics Data?
RNA-seq Tam on Acinetobacter baumannii strain ATCC 19606 CP059040.1 (Data_Tam_RNAseq_2024)
Deciphering S. aureus with metatranscriptomics (Marc's project)