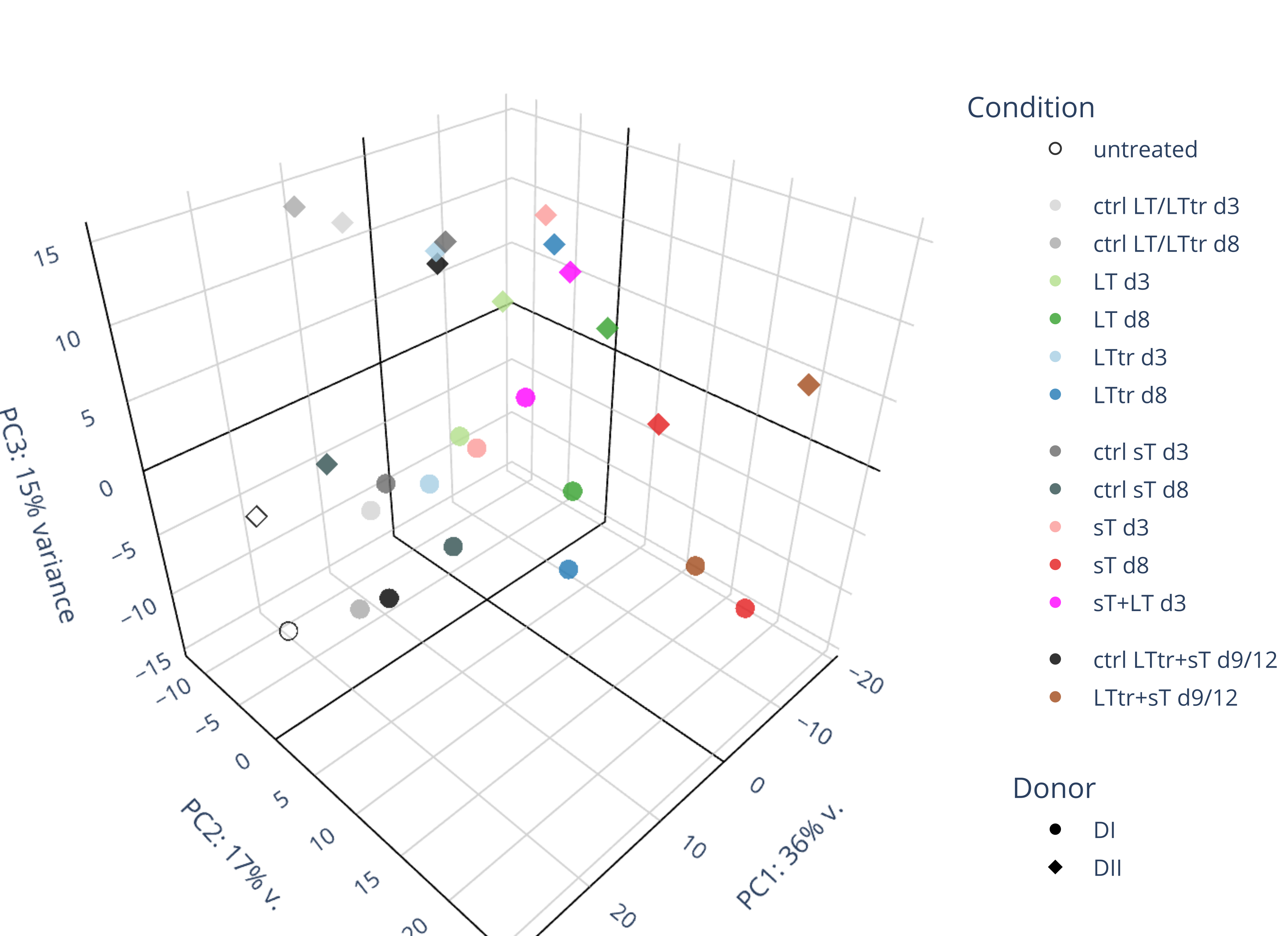
Draw 3D PCA and calculate background genes
gene_x 0 like s 1500 view s
Tags: plot, python
Download python3 script for generating the plot
Download raw data 1 for generating the plot
Download raw data 2 for generating the plot
construct a data structure (merged_df) as above with data and pc
library(ggplot2)
data <- plotPCA(rld, intgroup=c("condition", "donor"), returnData=TRUE)
write.csv(data, file="plotPCA_data.csv")
#calculate all PCs including PC3 with the following codes
library(genefilter)
ntop <- 500
rv <- rowVars(assay(rld))
select <- order(rv, decreasing = TRUE)[seq_len(min(ntop, length(rv)))]
mat <- t( assay(rld)[select, ] )
pc <- prcomp(mat)
pc$x[,1:3]
#df_pc <- data.frame(pc$x[,1:3])
df_pc <- data.frame(pc$x)
identical(rownames(data), rownames(df_pc)) #-->TRUE
## define the desired order of row names
#desired_order <- rownames(data)
## sort the data frame by the desired order of row names
#df <- df[match(desired_order, rownames(df_pc)), ]
data$PC1 <- NULL
data$PC2 <- NULL
merged_df <- merge(data, df_pc, by = "row.names")
#merged_df <- merged_df[, -1]
row.names(merged_df) <- merged_df$Row.names
merged_df$Row.names <- NULL # remove the "name" column
merged_df$name <- NULL
merged_df <- merged_df[, c("PC1","PC2","PC3","PC4","PC5","PC6","PC7","PC8","PC9","PC10","PC11","PC12","PC13","PC14","PC15","PC16","PC17","PC18","PC19","PC20","PC21","PC22","PC23","PC24","PC25","PC26","PC27","PC28","group","condition","donor")]
#results in 26PCs: merged_df <- merged_df[, c("PC1","PC2","PC3","PC4","PC5","PC6","PC7","PC8","PC9","PC10","PC11","PC12","PC13","PC14","PC15","PC16","PC17","PC18","PC19","PC20","PC21","PC22","PC23","PC24","PC25","PC26","group","condition","donor")]
write.csv(merged_df, file="merged_df_28PCs.csv")
#> summary(pc) #--> variances of PC1, PC2 and PC3 are 0.6795 0.08596 0.06599
#Importance of components:
# PC1 PC2 PC3 PC4 PC5 PC6 PC7
#Standard deviation 2.6011 0.92510 0.81059 0.67065 0.51952 0.46429 0.41632
#Proportion of Variance 0.6795 0.08596 0.06599 0.04517 0.02711 0.02165 0.01741
#Cumulative Proportion 0.6795 0.76548 0.83148 0.87665 0.90376 0.92541 0.94282
# PC8 PC9 PC10 PC11 PC12 PC13 PC14
#Standard deviation 0.38738 0.32048 0.27993 0.24977 0.20217 0.17316 0.16293
#Proportion of Variance 0.01507 0.01032 0.00787 0.00627 0.00411 0.00301 0.00267
#Cumulative Proportion 0.95789 0.96821 0.97608 0.98234 0.98645 0.98946 0.99213
# PC15 PC16 PC17 PC18 PC19 PC20 PC21
#Standard deviation 0.15058 0.13083 0.12449 0.08789 0.06933 0.06064 0.04999
#Proportion of Variance 0.00228 0.00172 0.00156 0.00078 0.00048 0.00037 0.00025
#Cumulative Proportion 0.99441 0.99612 0.99768 0.99846 0.99894 0.99931 0.99956
# PC22 PC23 PC24 PC25 PC26
#Standard deviation 0.04143 0.03876 0.03202 0.01054 0.005059
#Proportion of Variance 0.00017 0.00015 0.00010 0.00001 0.000000
#Cumulative Proportion 0.99973 0.99988 0.99999 1.00000 1.000000
draw 3D with merged_df using plot3D
import plotly.graph_objects as go
import pandas as pd
from sklearn.decomposition import PCA
import numpy as np
# Read in data as a pandas dataframe
#df = pd.DataFrame({
# 'PC1': [-13.999925, -12.504291, -12.443057, -13.065235, -17.316215],
# 'PC2': [-1.498823, -3.342411, -6.067055, -8.205809, 3.293993],
# 'PC3': [-3.335085, 15.207755, -14.725450, 15.078469, -6.917358],
# 'condition': ['GFP d3', 'GFP d3', 'GFP d8', 'GFP d8', 'GFP+mCh d9/12'],
# 'donor': ['DI', 'DII', 'DI', 'DII', 'DI']
#})
df = pd.read_csv('merged_df_28PCs.csv', index_col=0, header=0)
df['condition'] = df['condition'].replace("GFP+mCh d9/12", "ctrl LTtr+sT d9/12")
df['condition'] = df['condition'].replace("sT+LTtr d9/12", "LTtr+sT d9/12")
df['condition'] = df['condition'].replace("GFP d3", "ctrl LT/LTtr d3")
df['condition'] = df['condition'].replace("GFP d8", "ctrl LT/LTtr d8")
df['condition'] = df['condition'].replace("mCh d3", "ctrl sT d3")
df['condition'] = df['condition'].replace("mCh d8", "ctrl sT d8")
# Fit PCA model to reduce data dimensions to 3
pca = PCA(n_components=3)
pca.fit(df.iloc[:, :-3])
X_reduced = pca.transform(df.iloc[:, :-3])
# Add reduced data back to dataframe
df['PC1'] = X_reduced[:, 0]
df['PC2'] = X_reduced[:, 1]
df['PC3'] = X_reduced[:, 2]
# Create PCA plot with 3D scatter
fig = go.Figure()
#['circle', 'circle-open', 'square', 'square-open', 'diamond', 'diamond-open', 'cross', 'x']
# if donor == 'DI' else marker=dict(size=2, opacity=0.8, color=condition_color, symbol=donor_symbol)
#decrease diamond size to 6 while keep the circle as size 10 in the following code:
#'rgb(128, 150, 128)'
#I need three families of colors, always from light to deep, the first one should close to grey.
#the first serie for 'ctrl LTtr+sT d9/12', 'LTtr+sT d9/12'
#the second serie for 'ctrl LT/LTtr d3', 'ctrl LT/LTtr d8', 'LT d3', 'LT d8', 'LTtr d3', 'LTtr d8'
#the third serie for 'ctrl sT d3', 'ctrl sT d8', 'sT d3', 'sT d8', 'sT+LT d3'
condition_color_map_untreated = {'untreated':'black'}
donor_symbol_map_untreated = {'DI': 'circle-open', 'DII': 'diamond-open'}
#condition_color_map = {'ctrl LTtr+sT d9/12': 'green', 'GFP d3': 'blue', 'GFP d8': 'red', 'GFP+mCh d9/12': 'green', 'LT d3': 'orange'}
condition_color_map = {
'ctrl LTtr+sT d9/12': 'black',
'LTtr+sT d9/12': '#a14a1a',
'ctrl LT/LTtr d3': '#fdbf6f',
'ctrl LT/LTtr d8': '#ff7f00',
'LT d3': '#b2df8a',
'LT d8': '#33a02c',
'LTtr d3': '#a6cee3',
'LTtr d8': '#1f78b4',
'ctrl sT d3': 'rgb(200, 200, 200)',
'ctrl sT d8': 'rgb(100, 100, 100)',
'sT d3': '#fb9a99',
'sT d8': '#e31a1c',
'sT+LT d3': 'magenta'
}
donor_symbol_map = {'DI': 'circle', 'DII': 'diamond'}
for donor, donor_symbol in donor_symbol_map_untreated.items():
for condition, condition_color in condition_color_map_untreated.items():
mask = (df['condition'] == condition) & (df['donor'] == donor)
fig.add_trace(go.Scatter3d(x=df.loc[mask, 'PC1'], y=df.loc[mask, 'PC2'], z=df.loc[mask, 'PC3'],
mode='markers',
name=f'{condition}' if donor == 'DI' else None,
legendgroup=f'{condition}',
showlegend=True if donor == 'DI' else False,
marker=dict(size=6 if donor_symbol in ['diamond-open'] else 10, opacity=0.8, color=condition_color, symbol=donor_symbol)))
for donor, donor_symbol in donor_symbol_map.items():
for condition, condition_color in condition_color_map.items():
mask = (df['condition'] == condition) & (df['donor'] == donor)
fig.add_trace(go.Scatter3d(x=df.loc[mask, 'PC1'], y=df.loc[mask, 'PC2'], z=df.loc[mask, 'PC3'],
mode='markers',
name=f'{condition}' if donor == 'DI' else None,
legendgroup=f'{condition}',
showlegend=True if donor == 'DI' else False,
marker=dict(size=6 if donor_symbol in ['diamond'] else 10, opacity=0.8, color=condition_color, symbol=donor_symbol)))
for donor, donor_symbol in donor_symbol_map.items():
fig.add_trace(go.Scatter3d(x=[None], y=[None], z=[None],
mode='markers',
name=donor,
legendgroup=f'{donor}',
showlegend=True,
marker=dict(size=10, opacity=1, color='black', symbol=donor_symbol),
hoverinfo='none'))
# Annotations for the legend blocks
fig.update_layout(
annotations=[
dict(x=1.1, y=1.0, xref='paper', yref='paper', showarrow=False,
text='Condition', font=dict(size=15)),
dict(x=1.1, y=0.6, xref='paper', yref='paper', showarrow=False,
text='Donor', font=dict(size=15))
],
scene=dict(
aspectmode='cube',
xaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC1: 36% v.'),
yaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC2: 17% v.'),
zaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC3: 15% variance'),
bgcolor='white'
),
margin=dict(l=5, r=5, b=5, t=0) # Adjust the margins to prevent clipping of axis titles
)
#fig.show()
fig.write_image("fig1.svg")
calculate background genes
# [1] ensembl_gene_id gene_name MKL.1_RNA
# [4] MKL.1_RNA_118 MKL.1_RNA_147 MKL.1_EV.RNA
# [7] MKL.1_EV.RNA_2 MKL.1_EV.RNA_118 MKL.1_EV.RNA_87
#[10] MKL.1_EV.RNA_27 X042_MKL.1_wt_EV X0505_MKL.1_wt_EV
#[13] X042_MKL.1_sT_DMSO X0505_MKL.1_sT_DMSO_EV X042_MKL.1_scr_DMSO_EV
#[16] X0505_MKL.1_scr_DMSO_EV X042_MKL.1_sT_Dox X0505_MKL.1_sT_Dox_EV
#[19] X042_MKL.1_scr_Dox_EV X0505_MKL.1_scr_Dox_EV
"ensembl_gene_id
gene_name
python process.py ../fpkm_values_MKL-1.csv 3 > MKL.1_RNA
python process.py ../fpkm_values_MKL-1.csv 4 > MKL.1_RNA_118
python process.py ../fpkm_values_MKL-1.csv 5 > MKL.1_RNA_147
python process.py ../fpkm_values_MKL-1.csv 6 > MKL.1_EV.RNA
python process.py ../fpkm_values_MKL-1.csv 7 > MKL.1_EV.RNA_2
python process.py ../fpkm_values_MKL-1.csv 8 > MKL.1_EV.RNA_118
python process.py ../fpkm_values_MKL-1.csv 9 > MKL.1_EV.RNA_87
python process.py ../fpkm_values_MKL-1.csv 10 > MKL.1_EV.RNA_27
python process.py ../fpkm_values_MKL-1.csv 11 > X042_MKL.1_wt_EV
python process.py ../fpkm_values_MKL-1.csv 12 > X0505_MKL.1_wt_EV
python process.py ../fpkm_values_MKL-1.csv 13 > X042_MKL.1_sT_DMSO
python process.py ../fpkm_values_MKL-1.csv 14 > X0505_MKL.1_sT_DMSO_EV
python process.py ../fpkm_values_MKL-1.csv 15 > X042_MKL.1_scr_DMSO_EV
python process.py ../fpkm_values_MKL-1.csv 16 > X0505_MKL.1_scr_DMSO_EV
python process.py ../fpkm_values_MKL-1.csv 17 > X042_MKL.1_sT_Dox
python process.py ../fpkm_values_MKL-1.csv 18 > X0505_MKL.1_sT_Dox_EV
python process.py ../fpkm_values_MKL-1.csv 19 > X042_MKL.1_scr_Dox_EV
python process.py ../fpkm_values_MKL-1.csv 20 > X0505_MKL.1_scr_Dox_EV
python process.py ../fpkm_values_MKL-1.csv 21 > median_length
cut -f1 -d',' MKL.1_RNA>_MKL-1_RNA
cut -f1 -d',' MKL.1_RNA_118>_MKL-1_RNA_118
cut -f1 -d',' MKL.1_RNA_147>_MKL-1_RNA_147
cut -f1 -d',' MKL.1_EV.RNA>_MKL-1_EV_RNA
cut -f1 -d',' MKL.1_EV.RNA_2>_MKL-1_EV_RNA_2
cut -f1 -d',' MKL.1_EV.RNA_118>_MKL-1_EV_RNA_118
cut -f1 -d',' MKL.1_EV.RNA_87>_MKL-1_EV_RNA_87
cut -f1 -d',' MKL.1_EV.RNA_27>_MKL-1_EV_RNA_27
cut -f1 -d',' X042_MKL.1_wt_EV>_X042_MKL-1_wt_EV
cut -f1 -d',' X0505_MKL.1_wt_EV>_X0505_MKL-1_wt_EV
cut -f1 -d',' X042_MKL.1_sT_DMSO>_X042_MKL-1_sT_DMSO
cut -f1 -d',' X0505_MKL.1_sT_DMSO_EV>_X0505_MKL-1_sT_DMSO_EV
cut -f1 -d',' X042_MKL.1_scr_DMSO_EV>_X042_MKL-1_scr_DMSO_EV
cut -f1 -d',' X0505_MKL.1_scr_DMSO_EV>_X0505_MKL-1_scr_DMSO_EV
cut -f1 -d',' X042_MKL.1_sT_Dox>_X042_MKL-1_sT_Dox
cut -f1 -d',' X0505_MKL.1_sT_Dox_EV>_X0505_MKL-1_sT_Dox_EV
cut -f1 -d',' X042_MKL.1_scr_Dox_EV>_X042_MKL-1_scr_Dox_EV
cut -f1 -d',' X0505_MKL.1_scr_Dox_EV>_X0505_MKL-1_scr_Dox_EV
~/Tools/csv2xls-0.4/csv_to_xls.py _MKL-1_RNA _MKL-1_RNA_118 _MKL-1_RNA_147 _MKL-1_EV_RNA _MKL-1_EV_RNA_2 _MKL-1_EV_RNA_118 _MKL-1_EV_RNA_87 _MKL-1_EV_RNA_27 _X042_MKL-1_wt_EV _X0505_MKL-1_wt_EV _X042_MKL-1_sT_DMSO _X0505_MKL-1_sT_DMSO_EV _X042_MKL-1_scr_DMSO_EV _X0505_MKL-1_scr_DMSO_EV _X042_MKL-1_sT_Dox _X0505_MKL-1_sT_Dox_EV _X042_MKL-1_scr_Dox_EV _X0505_MKL-1_scr_Dox_EV -d',' -o background_genes_MKL-1.xls;
process.py
import sys
filename = sys.argv[1]
n = int(sys.argv[2])
with open(filename, 'r') as file:
for line in file:
words = line.split(",")
if len(words) >= n and words[n-1] != '0':
print(line.strip())
# -- in the example of K331A data --
#FORMAT normalized_counts.txt to normalized_counts.csv
#DEBUG1: add gene_symbol in the first line
#DEBUG2: '\n' --> ',\n'
#RESULT: it looks like #gene_symbol,ctrl_d3_DonorI,ctrl_d3_DonorII,ctrl_d8_DonorI,ctrl_d8_DonorII,LT_d3_DonorI,LT_d3_DonorII,LT_d8_DonorI,LT_d8_DonorII,LTtr_d3_DonorI,LTtr_d3_DonorII,LTtr_d8_DonorI,LTtr_d8_DonorII,K331A_DonorI,K331A_DonorII,
#DDX11L1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,
python process.py normalized_counts.csv 2 > ctrl_d3_DonorI
python process.py normalized_counts.csv 3 > ctrl_d3_DonorII
python process.py normalized_counts.csv 4 > ctrl_d8_DonorI
python process.py normalized_counts.csv 5 > ctrl_d8_DonorII
python process.py normalized_counts.csv 6 > LT_d3_DonorI
python process.py normalized_counts.csv 7 > LT_d3_DonorII
python process.py normalized_counts.csv 8 > LT_d8_DonorI
python process.py normalized_counts.csv 9 > LT_d8_DonorII
python process.py normalized_counts.csv 10 > LTtr_d3_DonorI
python process.py normalized_counts.csv 11 > LTtr_d3_DonorII
python process.py normalized_counts.csv 12 > LTtr_d8_DonorI
python process.py normalized_counts.csv 13 > LTtr_d8_DonorII
python process.py normalized_counts.csv 14 > K331A_DonorI
python process.py normalized_counts.csv 15 > K331A_DonorII
cut -f1 -d',' ctrl_d3_DonorI > _ctrl_d3_DonorI
cut -f1 -d',' ctrl_d3_DonorII > _ctrl_d3_DonorII
cut -f1 -d',' ctrl_d8_DonorI > _ctrl_d8_DonorI
cut -f1 -d',' ctrl_d8_DonorII > _ctrl_d8_DonorII
cut -f1 -d',' LT_d3_DonorI > _LT_d3_DonorI
cut -f1 -d',' LT_d3_DonorII > _LT_d3_DonorII
cut -f1 -d',' LT_d8_DonorI > _LT_d8_DonorI
cut -f1 -d',' LT_d8_DonorII > _LT_d8_DonorII
cut -f1 -d',' LTtr_d3_DonorI > _LTtr_d3_DonorI
cut -f1 -d',' LTtr_d3_DonorII > _LTtr_d3_DonorII
cut -f1 -d',' LTtr_d8_DonorI > _LTtr_d8_DonorI
cut -f1 -d',' LTtr_d8_DonorII > _LTtr_d8_DonorII
cut -f1 -d',' K331A_DonorI > _K331A_DonorI
cut -f1 -d',' K331A_DonorII > _K331A_DonorII
~/Tools/csv2xls-0.4/csv_to_xls.py _ctrl_d3_DonorI _ctrl_d3_DonorII _ctrl_d8_DonorI _ctrl_d8_DonorII _LT_d3_DonorI _LT_d3_DonorII _LT_d8_DonorI _LT_d8_DonorII _LTtr_d3_DonorI _LTtr_d3_DonorII _LTtr_d8_DonorI _LTtr_d8_DonorII _K331A_DonorI _K331A_DonorII -d',' -o background_genes.xls;
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Kraken2 Installation and Usage Guide
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Should the inputs for GSVA be normalized or raw?
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- Risks of Rebooting into Rescue Mode
- 足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
YopQ Secretion Boxplot and Fitting Function
RNA-seq skin organoids on GRCh38+chrHsv1 (final)
Genomic Organization of Herpes Simplex Virus type 1 (HSV-1 s17)